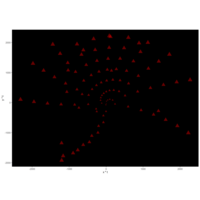
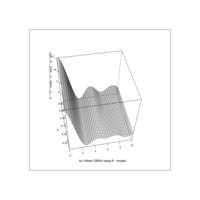
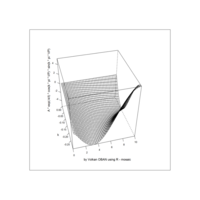
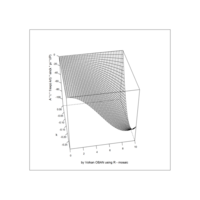
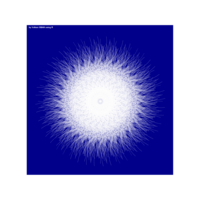
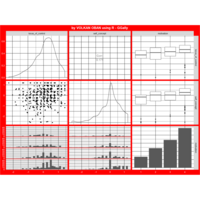
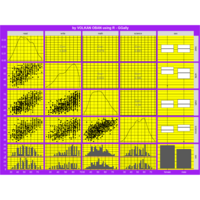
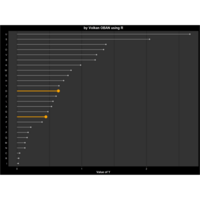
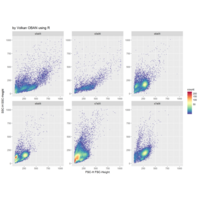
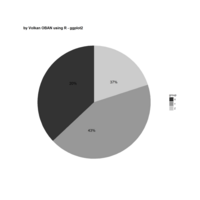
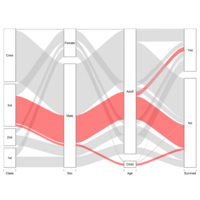
Recently Published

aRt with mathematics
function(x) : {x+tan(exp(sin(x)*cos(x-1)))}

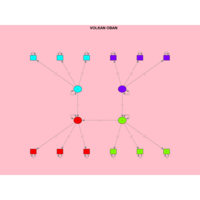
VOLKAN OBAN
R artsy package

R
VOLKAN OBAN
aRtsy

R
aRtsy

R
VOLKAN OBAN
aRtsy pckage

R
sin(x/tan(cos(x)))-exp(-sin(x))

R
ref: https://github.com/marcusvolz

aRt with mathematics
{tan(cos(x/x^3+3)/sin(x/x^3+1)-x^4)}

Plotting using complex functions
z^5+(-0.2+0.11*1i)/z^10
Plotting using complex functions
z^5+(-0.2+0.11*1i)/z^3

Plotting using complex functions
z^5+(-0.2+0.11*1i)/z^9

Plotting using complex functions
z^3+(-0.2+0.11*1i)/z^3

Plotting using complex functions
1+ z+z^2-0.8/z^3

VOLKAN OBAN
18,350,0.43,120,0.45,0.817,-0,12

art with mathematical functions
sin(cos(tan(exp(2-x))))

art with mathematical functions
12,250,0.41,110,0.25,1.817,-0.025
function(x) {cos(1/1+sin(x)+1/1+sin(x)*sin(x))}
aRt
sin(sin(cos((exp(1/x)/1+x^2)))/1+x^exp(-x^1/x^2))

art with mathematical functions
sin(cos(sin(x/1+x^4))/x+x^exp(-x^2/x))

aRt with mathematics
sin(sin(cos(x))/1+x^exp(-x^1/x))

aRt with mathematics
sin(x/1+x^exp(-x^1/x))
2.75
-0.25

aRt with mathematics
sin(1/1+x^exp(-x^1/x))

aRt
sin(exp(-1/cos(x/x^7+3)/x^9))

aRt with mathematics
sin(exp(-1/cos(x/x^7+3)/x^7))

aRt with mathematics
12
205
0.22
102 #
0.87 0.15
-0.28
{sin(cos(x/x^3+3)/sin(x/x^3+1)-x^4)}

aRt with mathematics
sin(cos(x+5*x*x/x^4+3)/sin(x/x^4+1)-x^3)

Plot
8
210
0.22
105
0.45
0.12
{sin(cos(x+5*x*x/3)/sin(x/x+1)-x^3)}

aRt with mathematics
sin(cos(2*sin(exp(x^sin(1/x^4)))))

art with mathematical functions
cos(2*sin(exp(x^sin(1/x^4))))

aRt with mathematics
cos(x*sin(exp(x^sin(1/x^2))))

art with mathematical functions
cos(x*sin(exp(x^sin(1/x^2)))

aRt with mathematics
cos(2*sin(exp(x^sin(x))))

Roses-art with mathematical functions
12
300
0.32
400
0.4
0.75
line_color <- "white"
back_color <- "black"
{sinh(log(x+1)*cos(x)*sin(1/x))}

R
{cos(sin(exp(-x^2))/x^3)}

aRt with mathematics
{cos(sin(exp(-x^2))/x^4)}

aRt with mathematics
cos(sin(exp(-x^2))/x^4)

aRt with mathematics
cos(x^4*x-sin(cos(1/x^4)))

art with mathematical functions
{sin(cos(1/x^4))}

aRt with mathematics
{sin(cos(1/x^4))}

R
sin(x^sin(cos(x)))
aRt
x^sin(cos(x))

R
1/x^4*sin(x)
R
{tan(sin(cos(exp(x/x^2+1))))}

R aRT
tan(sin(cos(exp(x/x^2+1))))

R aRT
Elif

VOLKAN OBAN
Dr. VOLKAN OBAN

art with mathematical functions
sin(cos(sin(x*x)))

aRt with mathematics
cos(exp(-x)*sin(exp(-x)))
R
cos(exp(-x)*sin(exp(-x)))

art with mathematical functions
12,100,0.41,105,2,0.2,-0.05, {sin(x/(-cos(x)))}

R
{sin(2*x*x)^x*x-x/cos(x+tan(x+1))}

R aRT
{sin(2*x*x)^x*x-x/cos(x+tan(x+1))}

art with mathematical functions
{sin(x)^x*x-1/cos(x^9)}
aRt with mathematics
{sin(x)^x*x-1/cos(x^9)}

mathematics
{sin(x)^x*x-1/cos(x^9)}

art with mathematical functions
{sin(x)^x-1/cos(x^9)}

VOLKAN OBAN
{sin(x)^2/cos(x*x)}

aRt with mathematics
{sin(cos(log(4^x*2)))/x^4-1}

aRt with mathematics
sin(cos(log(2^x*x+1)))/x^2-1

art with mathematical functions
{sin(cos(x/log(2^x*x+1)))+1}

art with mathematical functions
sin(cos(log(2^2*x+1)))/x^2-1)

art with mathematical functions
{{sin(cos(x+5*x*x/3)/sin(x/x+1)-x^3)}}

aRt with mathematics
{{sin(cos(x+5*x*x/3)/sin(x/x+1)-x^3)}}

R
Dr. Volkan OBAN

aRt with mathematics
{{cos(x/3)/sin(x/x*x+1)-x^5}}

R aRT
{cos(x/4)/1-x^5}

art with mathematical functions
{cos(x/2)/1-x^5}

aRt with mathematics
{cos(x/2)/1-x^3}

R
{x^x-sin(x^3)^tan(x/cos(x))}

VOLKAN OBAN
cos(x/x+x^(exp(-x*x)))

VOLKAN OBAN
{cos(x/x+x^2*(exp(-x*x)))}

aRt with mathematics
12 300 - 0.43 110 0.65 0.817 - -0.09 {cos(3*x/x+sin(exp(-x*x)))}

mathematical functions
{cos(2*x/x+sin(exp(-x*x)))}

aRt with mathematics
{cos(2*x/x+sin(exp(-x*x)))}
mathematical functions
sin(x+x^5/cos(sin(x)/x+2*sin(exp(-x))))

mathematical functions
{cos(x/2)/1-x^3}

aRt with mathematics
{sin(-cos(1/1x*x)*x/x^11+2)}

aRt with mathematics
{sin(cos(x)*x/x^5+2)}

aRt with mathematics
sin(cos(x)*1/x^3+2)

Plot
AÇI<- 2.15
cos(x/x^2^x/x^6+2)

aRt with mathematics
{cos(x/x^2^x/x^6+2)}
6 # 300 0.43 110 0.65 0.67 -0.09

mathematical functions
function(x) {cos(x/x^2^x/x^4+2)}

R
math
volkan oban
mathematics
sin(tan(abs(2*x)/x+1))

volkan oban
{cos(x)^3*x/x^2+1}
mathematics
{cos(2*sin(x/exp(-x))^1/x^2+1)}

aRt with mathematics
tan(x)*x+cos(x^7)

mathematics
log(cos(x^5))

volkan oban
cos(exp(-x))^sin(x^2)/x^7-1

aRt with mathematics
cos(exp(-x))^x/x^5-1

aRt with mathematics
cos(exp(-x))^x/x^3-1

art with mathematical functions
cos(2*sin(1/1+tan(exp(-*x))))

art with mathematical functions
cos(2*sin(x/exp(-x))^1/x+1)

mathematical functions
{sin(x*x+2/cos(exp(-x))^-x/x+1)}
mathematical functions
sin(cos

art with mathematical functions
exp(-sin(exp(cos(x/1-x^5)/x*x*x)))

art with mathematical functions
{exp(-sin(exp(cos(x)/x*x)))}

aRt with mathematics
5,250,0.43, 110,0.32,8.9-0.0002
{exp(-sin(exp(1/x*x)))}

aRt with mathematics
exp(-sin(exp(1/x*x)))

aRt with mathematics
12,101,0.43, 110,0.84,8.817,-0.0002
{exp(-sin(exp(1/x*x)))}

art with mathematical functions
sin(-exp(cos(-1/x*x*x)))

mathematical functions
cos(exp(sin(cos(exp(sin(cos(x)))))))

volkan oban
cos(exp(sin(cos(exp(sin(cos(x)))))))

math and graph
exp(exp(exp(exp(-x))))

aRt with mathematics
exp(sin(x))

mathematics
exp(x+log(sin(cos(sin(-exp(x*x))))))

mathematical functions
exp(-x)^log(sin(cos(sin(-exp(x*x)))))

volkan oban
x^log(sin(cos(sin(-exp(x*x)))))

art with mathematical functions
cos(sin(exp(x)))

aRt with mathematics
{cos(sin(exp(-x)))}

aRt with mathematics
8,1000,0.32,500,0.4,0.75,-0,27
cos(x)*sin(1/x)*log(x+1)

art with mathematical functions
sin(cos(sin(cos(x*x))))

Plot
12
> niter <- 200 #
> p <- 0.43 #
> st <- 48
>a lf <- 0.78 e
> aci <-2.817
> cv <- -0.05
> line_color<- "white"
> back_color <- "black"
function(x) {cos(exp(-x)*sin(2*x))}

art with mathematical functions
cos(2*tan(sin(-4*x*x*cos(tan(1/x*x*x*x)))))
aRt with mathematics
tan(1/exp(cos(4*x)))
aRt with mathematics
tan(exp(-cos(4*x)))
aRt with mathematics
tan(-exp(cos(x)))
Plot
tan(2*x)+cos(2*x)+sin(2*x)

volkan oban
abs(sin(cos(1/x*x))*exp(1/x*x))

aRt with mathematics
abs(sin(cos(1/x*x))*exp(-1/x))

art with mathematical functions
abs(sin(cos(1/x)))

art with mathematical functions
tan(exp(2*-cos(factorial(sin(x)))))
mathematics
x-factorial(sin(x))/x^2

mathematics
factorial(cos(x))

mathematics
tan(exp(2*-cos(factorial(sin(x)))))

aRt with mathematics
exp(sin(1/x))
12
250
0.4
101
0.25
-1.52

art with mathematical functions
exp(cos((sin(-x*x))))
aRt with mathematics
cos(x*x)*tan(x*x)*sin(x*x)*(sin(x))^2
aRt with mathematics
log(x+1)*tan(2*x)*sin(2*x)*(sin(x))^2
math and graph
sin(cos(exp(cos(1-x*x))))

aRt with mathematics
tan(x*x*exp(-sin(x)*cos(1/x/x)))

aRt with mathematics
x+tan(2*sin(exp(-sin(x))))

aRt with mathematics
{tan(2*sin(exp(-sin(x))))}
aRt with mathematics
tan(exp(sin(x)*cos(x)))

aRt with mathematics by Volkan OBAN
ref: Chinchón
aRt with mathematics
Trigonometric functions

aRt with mathematics
sin(exp(x)+cos(x))

aRt with mathematics
ref: A.S. Chinchón

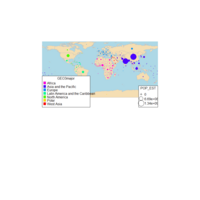
Johns Hopkins Covid-19 data
ref:https://joachim-gassen.github.io/
aRt with mathematics
{sin(1/cos(1+x))}

R volkan oban
{sin(-exp(cos(-1/x*x*x)))}

R
{sin(-exp(cos(-1/x*x*x)))}

aRt with mathematics
{cos(sinh(tan(-1/x)))+cosh(sin(x))}
R
{tan(sinh(x))}

aRt with mathematics
Volkan OBAN

aRt with mathematics
cos(sinh(tan(1/x)))

r volkan oban
function(x) {sinh((-cos(sin((1/x)+(1/x*x)+(1/x^3)+(1/x^4)+1))))}
R volkan oban
{sinh(cos((1/x)+(1/x*x)+(1/x^3)+(1/x^4)+1))}

R volkan oban
sinh((1/x)+(1/x*x)+(1/x^3)+(1/x^4))
aRt with mathematics
{1-cos(sinh(tan(sin(x))))/1-x}

R volkan oban
{x*cos(sinh(tan(sin(x))))/1-x}

aRt with mathematics
{cos(sinh(tan(sin(1-x))))}

aRt with mathematics
{cos(sinh(tan(sin(1-x))))}

aRt with mathematics
1-sinh(tan(sin(1-x)))

aRt with mathematics
{1-sinh(exp(-x))}

aRt with mathematics
{x+sinh(exp(-x))}

R volkan oban
sinh(cos(sin(exp(tan(cosh(x)/x*x)))))

R volkan oban
sinh(cos(sin(exp(tan(cosh(x)/x*x)))))

R volkan oban
{sin(exp(tan(-1/x*x)))

R volkan oban
{exp(tan(-1/x*x))}

R
{2*tan(1/x)-x}

R volkan oban
{2*x-x/cos(x)}
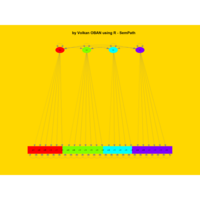
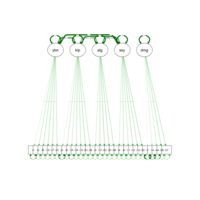
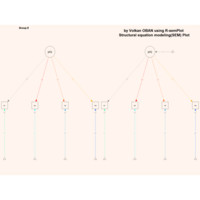
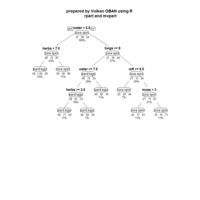
sem path
semPaths(fit,
+ sizeLat = 4, label.prop = 0.5, curve = 0.5, bg = "lightgreen", groups = "latents",
+ intercepts = FALSE, borders = FALSE, label.norm = "O")
> semPaths(fit,
+ sizeLat = 4, label.prop = 0.5, curve = 0.5, bg = "gold", groups = "latents",
+ intercepts = FALSE, borders = FALSE, label.norm = "O")
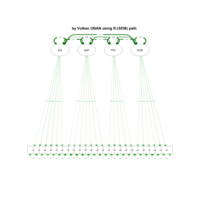
sem path
Structural Equation Modeling

aRt with mathematics
cos(1/x-exp(-4/x))

R volkan oban
{sin(sinh(x))}

aRt with mathematics
{cos(sin(x)-2*x)/x-log(x^5)}

R volkan oban
{cos(sin(x)-4*x)/x-log(x^5)}

aRt with mathematics
{cos(x)/x-log(x^5)}

aRt with mathematics
{1/x-log(x^3)

aRt with mathematics
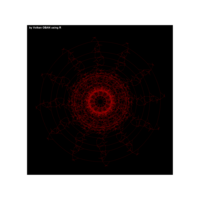
{sin(tan(exp(sin(x)*cos(x-1))))}
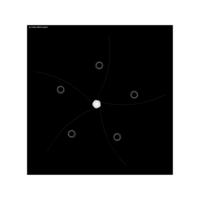
aRt with mathematics
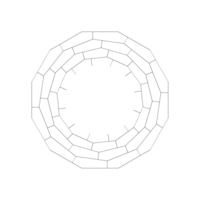
> edges <- 5 # Number of edges of the original polygon
> niter <- 300 # Number of iterations
> pond <- 0.43 # Weight to calculate the point on the middle of each edge
> step <- 101 # Number of times to draw mid-segments before connect ending points
> alph <- 0.25 # transparency of curves in geom_curve
> angle <- 0.817 # angle of mid-segment with the edge
> curv <- 0.197 # Curvature of curves
> line_color <- "white" # Color of curves in geom_curve
> back_color <- "black" # Background of the ggplot
> ratio_f <- function(x) {x+tan(exp(sin(x)*cos(x-1)))}

aRt
{x*(x+tan(exp(sin(x)*cos(x-1))))}

aRt with mathematics
{1/tan(1/exp(sin(cos(x))))+tan(cos(exp(-sin(x))))}

aRt with mathematics
VOLKAN OBAN

aRt with mathematics
tan(cos(exp(sin(x))))

aRt with mathematics
{sin(cos(exp(tan(x))))}

aRt with mathematics
function(x) {tan(sin(cos(1/x)))}

aRt with mathematics
{tan(sin(cos(x)))}
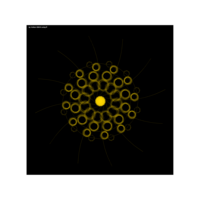
aRt with mathematics
function(x) {x+tan(exp(sin(x)*cos(x-1)))}

aRt with mathematics
function(x) {sin(x/4)}

aRt with mathematics
sin(x)/x-(cosh(exp(-sin(x))))}

aRt with mathematics
{sin(x)/x-(cosh(exp(-sin(x))))}

aRt with mathematics
{1/x-(cosh(exp(-cos(x))))}

aRt with mathematics
{1/x-(cosh(exp(-cos(x))))}

aRt with mathematics
1/x-(-sinh(exp(-cos(x))))

aRt with mathematics
{x-(-sinh(exp(-cos(x))))}

aRt with mathematics
1-(sinh(exp(cos(x))))

aRt with mathematics
{1-(-tan(exp(cos(x))))}

aRt with mathematics
{x/1-x-cos(x)*sin(tan(exp(cos(x/2))))}

flowers
log(x+1)*cos(x)*sin(1/x)

aRt with mathematics
-sin(x)*cos(x)*tan(x)

aRt with mathematics
function(x) {x/1-x-cos(x)*sin(-tan(exp(cos(x))))}

R
> library(tidyverse)
>
> # This function creates the segments of the original polygon
> polygon <- function(n) {
+ tibble(
+ x = accumulate(1:(n-1), ~.x+cos(.y*2*pi/n), .init = 0),
+ y = accumulate(1:(n-1), ~.x+sin(.y*2*pi/n), .init = 0),
+ xend = accumulate(2:n, ~.x+cos(.y*2*pi/n), .init = cos(2*pi/n)),
+ yend = accumulate(2:n, ~.x+sin(.y*2*pi/n), .init = sin(2*pi/n)))
+ }
>
> # This function creates segments from some mid-point of the edges
> mid_points <- function(d, p, a, i, FUN = ratio_f) {
+ d %>% mutate(
+ angle=atan2(yend-y, xend-x) + a,
+ radius=FUN(i),
+ x=p*x+(1-p)*xend,
+ y=p*y+(1-p)*yend,
+ xend=x+radius*cos(angle),
+ yend=y+radius*sin(angle)) %>%
+ select(x, y, xend, yend)
+ }
>
> # This function connect the ending points of mid-segments
> con_points <- function(d) {
+ d %>% mutate(
+ x=xend,
+ y=yend,
+ xend=lead(x, default=first(x)),
+ yend=lead(y, default=first(y))) %>%
+ select(x, y, xend, yend)
+ }
>
> edges <- 5 # Number of edges of the original polygon
> niter <-300 # Number of iterations
> pond <- 0.24 # Weight to calculate the point on the middle of each edge
> step <- 32 # Number of times to draw mid-segments before connect ending points
> alph <- 0.25 # transparency of curves in geom_curve
> angle <- 0.6 # angle of mid-segment with the edge
> curv <- 0.119 # Curvature of curves
> line_color <- "black" # Color of curves in geom_curve
> back_color <- "white" # Background of the ggplot
> ratio_f <- function(x) {1/sin(x)} # To calculate the longitude of mid-segments
>
> # Generation on the fly of the dataset
> accumulate(.f = function(old, y) {
+ if (y%%step!=0) mid_points(old, pond, angle, y) else con_points(old)
+ }, 1:niter,
+ .init=polygon(edges)) %>% bind_rows() -> df
>
> # Plot
> ggplot(df)+
+ geom_curve(aes(x=x, y=y, xend=xend, yend=yend),
+ curvature = curv,
+ color=line_color,
+ alpha=alph)+
+ coord_equal()+
+ theme(legend.position = "none",
+ panel.background = element_rect(fill=back_color),
+ plot.background = element_rect(fill=back_color),
+ axis.ticks = element_blank(),
+ panel.grid = element_blank(),
+ axis.title = element_blank(),
+ axis.text = element_blank())
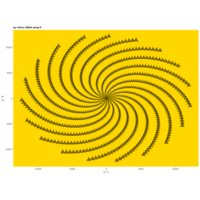
aRt
> angle <- 6.2
> points <- 1000
>
> t <- (1:points)*2*angle
> x <- sin(-2*t)
> y <- cos(2*t)
>
> df <- data.frame(t, x, y)
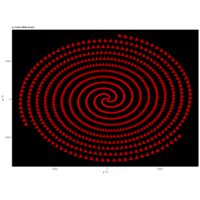
R
> angle <- 4.2
> points <- 1000
>
> t <- (1:points)*2*angle
> x <-cos(t)
> y <-sin(t)
>
> df <- data.frame(t, x, y)
>
R
> angle <- 4.2
> points <- 1000
>
> t <- (1:points)*angle
> x <-sin(t)
> y <- cos(t)*(-1/t)
>
> df <- data.frame(t, x, y)
R
> angle <- 4.2
> points <- 1000
>
> t <- (1:points)*angle
> x <- t-exp(-1/t)
> y <- cos(1/t)-sin(t)
>
> df <- data.frame(t, x, y)
>
> p <- ggplot(df, aes(x*t, y*t))
> p + geom_point(aes(size = t), alpha = 0.72, color = "red", shape = 17) +theme(
+ plot.title = element_text(color = "black", size = 7, face = "bold"),
+ panel.grid = element_blank(),
+ legend.position = "none",
+ panel.background = element_rect(fill = "black"))
R
> angle <- 4.2
> points <- 1000
>
> t <- (1:points)*angle
> x <- t
> y <- cos(1/t-t)
>
> df <- data.frame(t, x, y)
>
R DataViz
> angle <- 3.2
> points <- 1000
>
> t <- (1:points)*angle
> x <- sin(t^3-t^2+t)
> y <- cos(1/t-t)
>
> df <- data.frame(t, x, y)
R
> angle <- 3.2
> points <- 600
>
> t <- (1:points)*angle
> x <- sin(t^3-t)
> y <- cos(1/t)
>
> df <- data.frame(t, x, y)
R DataViz
> angle <- 4.2
> points <- 1000
>
> t <- (1:points)*2*angle
> x <- sin(tan(2*t))
> y <- cos(tan(2*t))
>
R
> angle <- 4.2
> points <- 1000
>
> t <- (1:points)*2*angle
> x <- sin(2*t)
> y <- cos(2*t)
>
> df <- data.frame(t, x, y)

aRt with mathematics
function(x) {cos(x+x^3+x^7)-sin(x)}
mosaic
exp
> library(manipulate)
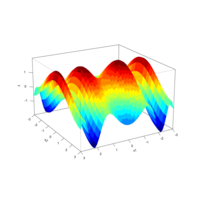
> plotFun(A *exp(-1/t)* cos(k*pi * t/P) * sin(2 * pi * t/P) ~ t + k, t.lim = range(0, 10),k.lim = range(-0.3,0), A = 10, P = 4, surface = TRUE)

aRt with mathematics
x*sin(x)-log(x)*cos(x)+1

aRt with mathematics
1-log(x)*[cos(x)*sin(x)*tan(x)/exp(x*x*x)]

aRt with mathematics
sin(x)+tan(x)/exp(x)

aRt with mathematics
1+cos(2*x)*log(x)*sin(x)

aRt with mathematics
1+x*log(x)*sin(x)

mathematical art
1-tan(2x)
aRt with mathematics
function(x) {x^2 -1 /x*sin(cos(sin(x)))*log(x+1)}

aRt with mathematics
function(x) {sin(x)/x*x}

aRt with mathematics
function(x) {(log(x+(x^2))*cos(sqrt(x))/exp((x^2)-1))+sin(1+x^3)+1-1/1-x}

aRt with mathematics
function(x) {(log(x+sqrt(x^5))*cos(sqrt(x))/exp((x^2)-1))+sin(1+x)+1+cos(x)
aRt with mathematics
• function(x) {(log(x+sqrt(x))*cos(x)/exp((x^3)-1))+sin(1+x)+1}

aRt with mathematics
• function(x) {(log(x)*cos(x)/exp((x^3)-1))+sin(1+x)+1}

aRt with mathematics
function(x) {(log(x)/exp((x^3)-1))+sin(1+x)}

aRt with mathematics
function(x) {(1/exp((x^3)-1))+sin(1+x)}

R
function(x) {x^3+sin(2*x)*cos(3/x)*log(2*x)+1/x-5*x}
ref:aschinchon

aRt
function(x) {x+cos(x*x-1)*sin(x*x-1)+(x-1)}

aRt with mathematics
function(x) {exp(cos(x*x-1))*sin(x*x*x)}

aRt with mathematics
function(x) {exp(cos(x*x-1))}

aRt with mathematics
function(x) {cos(x+1)*sin(x-1)-1/x-log(x)}

aRt with mathematics
function(x) {cos(x)*sin(x-1)-x*tan(1/x)+log(x)}

aRt with mathematics
function(x) {cos(x)*sin(x-1)-x}

R
function(x) {1/tan(-cos(sin(log(x*x/exp(-x^2)))))}

R
{tan(cos(sin(log(x*x/exp(-x^2))))}

R
function(x) {cos(sin(log10(x*x/500))/x}

R
function(x) {sin(log10(x*x/500))}
aRt with mathematics
log(5*x+1)*cos(3*x)*sin(1/x)
ggparty
ref: https://cran.r-project.org/web/packages/ggparty/vignettes/ggparty-graphic-partying.html

ggparty
ref:https://cran.r-project.org/web/packages/ggparty/vignettes/ggparty-graphic-partying.html

geometric shape
ref:Antonio Sánchez Chinchón
ggforce
ref: r-blogger
ggforce
ref: r- blogger
ggforce
ref : r blogger

network visualization
Network visualization in R.
library(igraph)
library(ggraph)
library(igraphdata)
library(smglr)
data: yeast
yeast protein interactions from igraphdata (only biggest component)
ref:https://lnkd.in/gasiqWz

chaos
ref: fronkonstin

chaos
ref:fronkonstin.com/category/chaos/

ggstatsplot
ggstatsplot

ggstatsplot
Package ‘ggstatsplot in R.
it supports only the most common types
of statistical tests: parametric, nonparametric, robust, and bayesian versions of t-test/anova, correlation analyses, contingency table analysis ,and regression analyses.
#R #volkanoban #statisticaltests #datascience #analytics #datavisualization
ref: cran.r-project.org

ggplot2
library(tidyverse)
> seq(from=-10, to=10, by = 0.05) %>%
+ expand.grid(x=., y=.) %>%
+ ggplot(aes(x=(x^2+0.5*pi*cos(y)^2), y=(y+0.5*pi*sin(x)))) +
+ geom_point(alpha=.1, shape=20, size=1, color="white")+
+ theme_void()+coord_fixed()
Plot
> theme <- theme(plot.title = element_text(hjust = 0.5), # Centered title
+ plot.background = element_rect(fill="blueviolet"), # Black background
+ panel.background = element_rect(fill="purple"), # Dark grey panel background
+ panel.grid.minor = element_line(color="blueviolet"), # Hide grid lines
+ panel.grid.major = element_line(color="blueviolet"), # Hide grid lines
+ axis.text = element_text(color="white"), # Make axis text white
+ title = element_text(color="white", face="bold"), # Make title white and bold.
+ legend.background = element_rect(fill="blueviolet"), # Make legend background black
+ legend.text = element_text(color="white"), # Make legend text white
+ legend.key = element_rect(fill="blueviolet", color="blueviolet"), #Squares/borders of legend black
+ legend.position = c(.9,.4)) # Coordinates. Top right = (1,1)
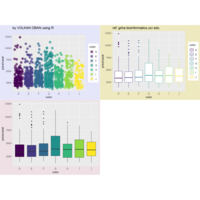
> ggplot(diamonds, aes(x=cut, y=price)) +
+ geom_boxplot(aes(color=clarity), fill=NA) +
+ scale_color_discrete(guide=F) +
+ facet_wrap(~clarity, ncol=2) + theme
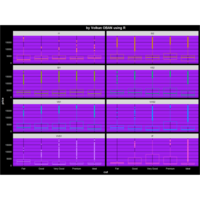
ggplot2
thm <- theme(plot.title = element_text(hjust = 0.5), # Centered title
+ plot.background = element_rect(fill="black"), # Black background
+ panel.background = element_rect(fill="purple"), #
+ panel.grid.minor = element_line(color="black"), # Hide grid lines
+ panel.grid.major = element_line(color="black"), # Hide grid lines
+ axis.text = element_text(color="white"), # Make axis text white
+ title = element_text(color="white", face="bold"), # Make title white and bold.
+ legend.background = element_rect(fill="black"), # Make legend background black
+ legend.text = element_text(color="white"), # Make legend text white
+ legend.key = element_rect(fill="black", color="black")

DALEX
breakDown::HR_data

factoextra
fviz_silhouett

k-means Clustering
factoextra and clustering packages
grid,gridextra
ref:https://uc-r.github.io/kmeans_clustering
Plot
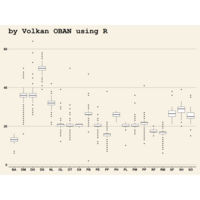
ggplot(surveys_complete, aes(x = species_id, y = hindfoot_length)) +
+ geom_boxplot() +
+ theme_wsj()

ggplot2
> ggplot(data = surveys_complete, mapping = aes(x = species_id, y = weight)) +
+ geom_boxplot(alpha = 0) +
+ geom_jitter(alpha = 0.3, color = "red")
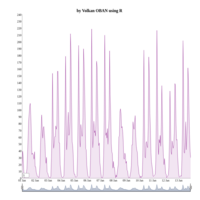
dygraphs
ref: r-graph-gallery

Calendar Heatmap
ref: r-graph-gallery.com

Calendar Heatmap
ref:
r-graph-gallery

Calendar Heatmap
ref:r-graph-gallery

wordcloud2 package
wordcloud2(d, size =1 , minRotation = -pi/8, maxRotation = -pi/3, rotateRatio = 1)

wordcloud2 package
Happy new years

ggwordcloud
love….AŞK

network visualization
ref: data-to-viz.com

network visualization
ref: data-to-viz.com
Plot
library(network)
library(sna)
library(maps)
library(ggplot2)

aRt
ref:fronkonstin.com

aRt
ref : fronkonstin.com

Cannibus Curve
,ref: r-bloggers.com/cannibus-curve-with-ggplot2/

lime
ref:www.data-imaginist.com

factoextra NbClust
ref : http://www.sthda.com

factoextra NbClust
ref :http://www.sthda.com

factoextra NbClust
ref: http://www.sthda.com
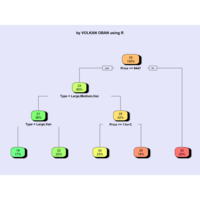
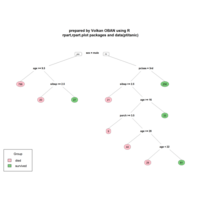
rpart.plot
> par(bg='lavender')
> anova.model <- rpart(Mileage ~ ., data = cu.summary)
> rpart.plot(anova.model, box.palette = "GnYlRd",
+ shadow.col = "black",
)

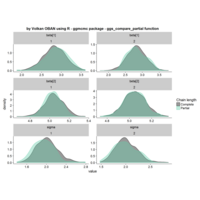
stacked densities plot
ref : shinyapps.
Michael Lee

Plot
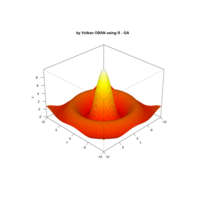
> par(bg='springgreen4')
> x <- seq(-10, 10, length = 80)
> y <- x
> f <- function(x, y) {r <- sqrt(x^2 + y^2); 10 * cos(2*r) / 2*r}
> z <- outer(x, y, f)
> persp(x, y, z,col='royalblue1')

ggplot2 and ggthemes
facet_wrap

Plot
v=2*pi*(3-sqrt(5))
> i=500
> ggplot(data.frame(r=sqrt(1:i),t=(1:n)*v),
+ aes(x=r*cos(t),y=r*sin(t)))+
+ geom_point(aes(x=0,y=0),
+ size=240,
+ colour="violetred")+
+ geom_point(aes(size=(n-r)),
+ shape=21,fill="black",
+ colour="purple")+
+ theme_void()+theme(legend.position="none")

Plot
ggplot(df, aes(x,y)) +
+ geom_polygon()+
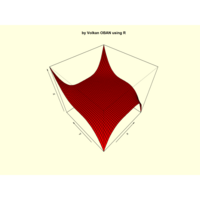
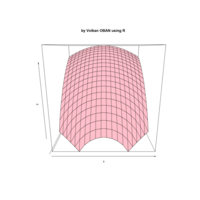
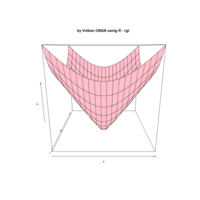
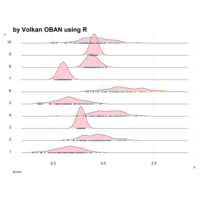
+ theme_void() + ggtitle("by VOLKAN OBAN using R \n Data Scientist")
> d <- data.frame(x=3, y=3)
> for (i in 2:1000){
+ d[i,1] <- d[i-1,1]+((0.88)^i)*2*cos(2*i)
+ d[i,2] <- d[i-1,2]+((0.88)^i)*2*sin(2*i)
+ }
> ggplot(df, aes(x,y)) +
+ geom_polygon()

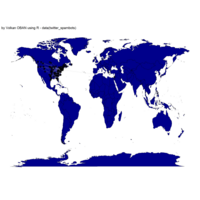
ggplot2
library(ggplot2)
library(grid)
# get data
download.file(url="http://www.naturalearthdata.com/http//www.naturalearthdata.com/download/110m/cultural/ne_110m_admin_0_countries.zip", "ne_110m_admin_0_countries.zip", "auto")
unzip("ne_110m_admin_0_countries.zip")
file.remove("ne_110m_admin_0_countries.zip")
# read shape file using rgdal library
library(rgdal)
ogrInfo(".", "ne_110m_admin_0_countries")
world <- readOGR(".", "ne_110m_admin_0_countries")
summary(world)
plot(world, col = "firebrick1")

art
ref: https://github.com/aschinchon

aRt
> seq(-3,3,by=.01) %>%
+ expand.grid(x=., y=.) %>%
+ ggplot(aes(x=(x^5-sin(y^2)), y=(y^5-cos(x^2)))) +
+ geom_point(alpha=.05, shape=20, size=0, color="white")+
+ theme_void()+
+ coord_fixed()+
+ theme(panel.background = element_rect(fill="darkred"))+
+ coord_polar()

aRt
library(tidyverse)
> seq(-3,3,by=.01) %>%
+ expand.grid(x=., y=.) %>%
+ ggplot(aes(x=(x^3-sin(y^2)), y=(y^3-cos(x^2)))) +
+ geom_point(alpha=.1, shape=20, size=0, color="white")+
+ theme_void()+
+ coord_fixed()+
+ theme(panel.background = element_rect(fill="purple"))+
+ coord_polar()
ref:
https://fronkonstin.com/

aRt
ref:https://fronkonstin.com/

aRt
df <- data.frame(x=0, y=0)
> for (i in 2:500){
+ df[i,1] <- df[i-1,1]+((0.98)^i)*cos(3*i)
+ df[i,2] <- df[i-1,2]+((0.98)^i)*sin(3*i)

aRt
ref: https://fronkonstin.com/2017/12/23/tiny-art-in-less-than-280-characters/
aRt
> t=seq(1, 80, by=.001)
> plot(exp(-0.005*t)*sin(t*3.019+2.677)+
+ exp(-0.001*t)*sin(t*2.959+2.719),
+ exp(-0.005*t)*sin(t*2.964+0.229)+
+ exp(-0.008*t)*sin(t*2.984+1.284),
+ type="l", axes=FALSE," , ylab="")
ggpubr
ggdonutchart
ggsci
theme(plot.background = element_rect(fill = "palegoldenrod"))

ggiraph
ref:r-graph-gallery.com

ggplot2 ggthemes pack.
> ggplot(dt.long,aes(factor(variable), value))+
+ geom_violin(aes(fill=factor(variable)))+
+ geom_boxplot(alpha=0.2, color="purple", width=.2)+
+ labs(x = "", y = "")+
+ theme_bw()+
+ theme(legend.title = element_blank())+
+ facet_wrap(~variable, scales="free")
ref: aledemogr.com

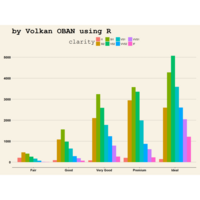
ggplot2
ggplot(diamonds, aes(cut)) +
+ geom_bar(aes(fill = clarity), position = "dodge") +
+ scale_fill_brewer(palette="PuOr") +
+ geom_hline(yintercept = 2710, color="black") +
+ annotate("text", x = 1.5, y=2250, label = "Threshold value", color= "darkred")

VOLKAN OBAN
Plotrix
Test color legends
Plotrix
clock24.plot
ref:https://cran.r-project.org/web/packages/plotrix/plotrix.pdf

Plotrix
ref:https://cran.r-project.org/web/packages/plotrix/plotrix.pdf

Plotrix
ref:https://cran.r-project.org/web/packages/plotrix/plotrix.pdf

"TSP" - The Travelling Salesman Problem (TSP).
ref:https://github.com/aschinchon/

spatstat
delaunay

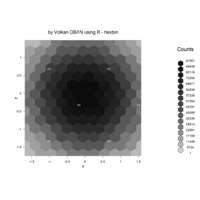
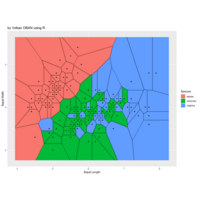
aRt with R
iter=5
> points=12 # Number of points
> radius=2.4
> angles=seq(0, 5*pi*(3-1/points), length.out = points)+pi/2
>
> df=data.frame(x=4, y=4)
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ { data.frame(x=df[i,"x"]+radius^(k-1)*cos(3*angles),
+ y=df[i,"y"]+radius^(k-1)*sin(3*angles)) %>% rbind(temp) -> temp
+
+ }
+ df=temp
+ }
> df %>%
+ select(x,y) %>%
+ deldir(sort=TRUE) %>%
+ .$dirsgs -> data
>
> # Plot regions with geom_segmen
> data %>%
+ ggplot() +
+ geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2), color="black") +
+ scale_x_continuous(expand=c(0,0))+
+ scale_y_continuous(expand=c(0,0))+
+ coord_fixed() +
+ theme(legend.position = "none",
+ panel.background = element_rect(fill="white"),
+ panel.border = element_rect(colour = "black", fill=NA),
+ axis.ticks = element_blank(),
+ panel.grid = element_blank(),
+ axis.title = element_blank(),
+ axis.text = element_blank())->plot
>
> plot

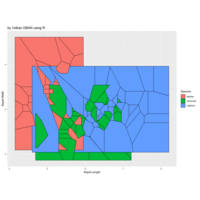
spatstat
delaunay

spatstat
dirichlet

mosaic
data:happy
ggmosaic package
NHANES
ggplot(data = NHANES) +
+ geom_mosaic(aes(weight = Weight, x = product(Age), fill=factor(SleepHrsNight)), na.rm=TRUE) + theme(axis.text.x=element_text(angle=0, hjust= .5))+labs(x="Age", y=" ggmosaic") + guides(fill=guide_legend(title = "SleepHrsNight", reverse = TRUE))

Plot
> library(ggplot2)
> library(dplyr)
> library(deldir)
>
> iter=4 # Number of iterations (depth)
> points=4# Number of points
> radius=2.4
> angles=seq(0, 4*pi*(5-1/points), length.out = points)+pi/2
>
> # Initial center
> df=data.frame(x=4, y=4)
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+radius^(2*k-2)*cos(5*angles),
+ y=2*df[i,"y"]+radius^(2*k-2)*sin(angles)) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }
> df %>%
+ select(x,y) %>%
+ deldir(sort=TRUE) %>%
+ .$dirsgs -> data
>
> data %>%
+ ggplot() ++
+ geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2), color="darkblue") +
+ scale_x_continuous(expand=c(0,0))+
+ scale_y_continuous(expand=c(0,0))+
+ coord_fixed() +
+ theme(legend.position = "none",
+ panel.background = element_rect(fill="magenta"),
+ panel.border = element_rect(colour = "black", fill=NA),
+ axis.ticks = element_blank(),
+ panel.grid = element_blank(),
+ axis.title = element_blank(),
+ axis.text = element_blank())->plot
>
> plot
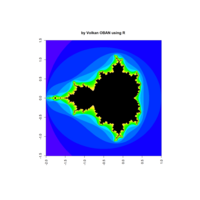
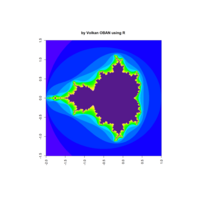
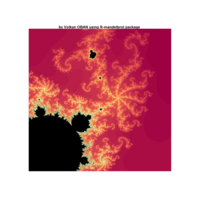
fractal-mandelbrot
z <- mandelbrot(iter=15)
> par(pty="s")
> image(z,col=c(topo.colors(n+6),"black"), las=3)
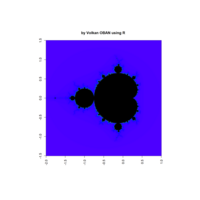
Plot
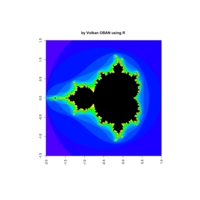
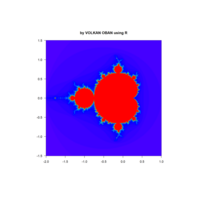
z <- mandelbrot(iter=400)
> par(pty="s")
> image(z,col=c(topo.colors(n+3),"black"), las=3)
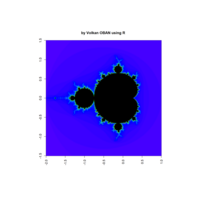
mandelbrot
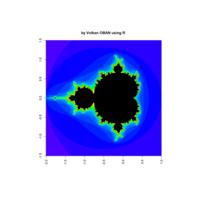
z <- mandelbrot(iter=100)
> par(pty="s")
> image(z, col=c(topo.colors(n+1),"black"), las=3)
ref:https://github.com/mariodosreis/fractal
fractal
library(fractal)
> z <- mandelbrot(iter=100)
> par(pty="s")
> image(z, col=c(topo.colors(n),"red"), las=1)

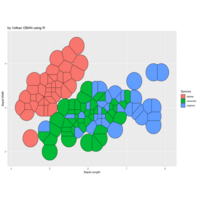
art game with R
iter=4
> points=16
> radius=4
> angles=seq(0, 18*pi*(3-1/points), length.out = points)+pi/2
> df=data.frame(x=7, y=7)
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+2*radius^(k-4)*cos(5*angles),
+ y=df[i,"y"]+2*radius^(k-4)*sin(5*angles)) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }
> df %>%
+ select(x,y) %>%
+ deldir(sort=TRUE) %>%
+ .$dirsgs -> data
> data %>%
+ geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2), color="magenta4") +
+ scale_x_continuous(expand=c(0,0))+
+ scale_y_continuous(expand=c(0,0))+
+ coord_fixed() +
+ theme(legend.position = "none",
+ panel.background = element_rect(fill="midnightblue"),
+ panel.border = element_rect(colour = "black", fill=NA),
+ axis.ticks = element_blank(),
+ panel.grid = element_blank(),
+ axis.title = element_blank(),
+ axis.text = element_blank())->plot
>

aRt with R
iter=4
> points=8
> radius=4
> angles=seq(0, 18*pi*(5-1/points), length.out = points)+pi/2
> df=data.frame(x=4, y=4)
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+2*radius^(k-3)*cos(3*angles),
+ y=df[i,"y"]+2*radius^(k-3)*sin(3*angles)) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }
>

art game with R
iter=4
> points=8
> radius=4
>
> angles=seq(0, 18*pi*(5-1/points), length.out = points)+pi/2
> df=data.frame(x=2, y=2)
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+2*radius^(k-3)*cos(2*angles),
+ y=df[i,"y"]+2*radius^(k-3)*sin(2*angles)) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }

aRt with R
iter=4
> points=8
> radius=4
> angles=seq(0, 18*pi*(5-1/points), length.out = points)+pi/2
>
> df=data.frame(x=1, y=1)
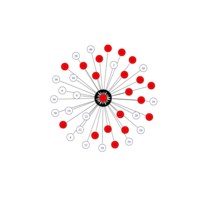
igraph and visNetwork
> g <- graph.star(40)
> V(g)$color <- c("red", "white")
>
> E(g)$color <- "black"
> plot(g)

visNetwork
nnodes <- 300
> nnedges <- 1500
> nodes <- data.frame(id = 1:nnodes)
> edges <- data.frame(from = sample(1:nnodes, nnedges, replace = T),
+ to = sample(1:nnodes, nnedges, replace = T))
> # with defaut layout
> visNetwork(nodes,edges) %>%
+ visIgraphLayout()
> # use full space
> visNetwork(nodes, edges") %>%
+ visIgraphLayout(type = "full")
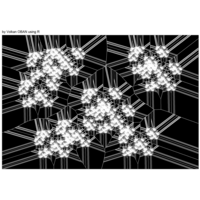
data aRt with R
...............
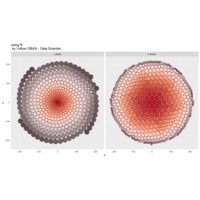
> iter=5 # Number of iterations (depth)
> points=10 # Number of points
> radius=4 # Factor of expansion/compression
>
> # Angles of points from center
> angles=seq(0, 12*pi*(5-1/points), length.out = points)+pi/2
>
> # Initial center
> df=data.frame(x=0, y=0)
>
> # Iterate over centers again and again
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+6*radius^(k-2)*cos(angles),
+ y=df[i,"y"]+4*radius^(k-2)*sin(2*angles)) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }
>
> # Obtain Voronoi regions
> df %>%
+ select(x,y) %>%
+ deldir(sort=TRUE) %>%
+ .$dirsgs -> data
>
> # Plot regions with geom_segmen
> data %>%
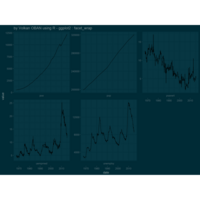
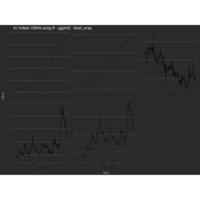
+ ggplot() + ggtitle("by Volkan OBAN using R ") +
+ geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2), color="white") +
+ scale_x_continuous(expand=c(0,0))+
+ scale_y_continuous(expand=c(0,0))+
+ coord_fixed() +
+ theme(legend.position = "none",
+ panel.background = element_rect(fill="black"),
+ panel.border = element_rect(colour = "black", fill=NA),
+ axis.ticks = element_blank(),
+ panel.grid = element_blank(),
+ axis.title = element_blank(),
+ axis.text = element_blank())->plot
................

Plot
............................................
> iter=5
> points=16 # Number of points
> radius=2.5 # Factor of expansion/compression
>
> # Angles of points from center
> angles=seq(0, 2*pi*(8-1/points), length.out = points)+pi/2
>
> # Initial center
> df=data.frame(x=0, y=0)
>
> # Iterate over centers again and again
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+3*radius^(k-1)*cos(angles),
+ y=df[i,"y"]+2*radius^(k-1)*sin(2*angles)) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }
...........................
.........

data aRt with R
data.frame(x=df[i,"x"]+radius^(k-1)*cos(angles^2),
+ y=df[i,"y"]+radius^(k-1)*sin(angles^2)) %>% rbind(temp) -> temp
colors: midnightblue and mediumpurple1

Plot
code:
ref:https://github.com/aschinchon/mandalas
library(dplyr)
> library(deldir)
>
> # Parameters to change as you like
> iter=5 # Number of iterations (depth)
> points=9 # Number of points
> radius=3.9 # Factor of expansion/compression
>
> # Angles of points from center
> angles=seq(0, 3*pi*(4-1/points), length.out = points)+pi/2
>
> # Initial center
> df=data.frame(x=0, y=0)
>
> # Iterate over centers again and again
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+radius^(k-1)*cos(angles),
+ y=df[i,"y"]+radius^(k-1)*sin(angles)) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }
>
> # Obtain Voronoi regions
> df %>%
+ select(x,y) %>%
+ deldir(sort=TRUE) %>%
+ .$dirsgs -> data
>
> # Plot regions with geom_segmen
> data %>%
+ ggplot()
+ geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2), color="white") +
+ scale_x_continuous(expand=c(0,0))+
+ scale_y_continuous(expand=c(0,0))+
+ coord_fixed() +
+ theme(legend.position = "none",
+ panel.background = element_rect(fill="black"),
+ panel.border = element_rect(colour = "white", fill=NA),
+ axis.ticks = element_blank(),
+ panel.grid = element_blank(),
+ axis.title = element_blank(),
+ axis.text = element_blank())->plot
>
> plot

data aRt with R -Mandalas
> iter=4 # Number of iterations (depth)
> points=8 # Number of points
> radius=4 # Factor of expansion/compression
>
> # Angles of points from center
> angles=seq(0, 2*pi*(30-1/points), length.out = points)+pi/2
>
> # Initial center
> df=data.frame(x=0, y=0)
>
> # Iterate over centers again and again
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+radius^(k-1)*cos(angles/4),
+ y=df[i,"y"]+radius^(k-1)*sin(angles/4)) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }

data visulazition in R
> library(ggplot2)
> library(dplyr)
> library(deldir)
>
> # Parameters to change as you like
> iter=4 # Number of iterations (depth)
> points=8 # Number of points
> radius=4 # Factor of expansion/compression
>
> # Angles of points from center
> angles=seq(0, 2*pi*(20-1/points), length.out = points)+pi/2
>
> # Initial center
> df=data.frame(x=0, y=0)
>
> # Iterate over centers again and again
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+radius^(k-1)*cos(angles/2),
+ y=df[i,"y"]+radius^(k-1)*sin(angles/2)) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }
>
> # Obtain Voronoi regions
> df %>%
+ select(x,y) %>%
+ deldir(sort=TRUE) %>%
+ .$dirsgs -> data
>
> # Plot regions with geom_segmen
> data %>%
+ ggplot()
+ geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2), color="purple4") +
+ scale_x_continuous(expand=c(0,0))+
+ scale_y_continuous(expand=c(0,0))+
+ coord_fixed() +
+ theme(legend.position = "none",
+ panel.background = element_rect(fill="plum2"),
+ panel.border = element_rect(colour = "black", fill=NA),
+ axis.ticks = element_blank(),
+ panel.grid = element_blank(),
+ axis.title = element_blank(),
+ axis.text = element_blank())->plot
>
> plot

DATA ART with R
> library(ggplot2)
> library(dplyr)
> library(deldir)
>
> # Parameters to change as you like
> iter=4 # Number of iterations (depth)
> points=8 # Number of points
> radius=4 # Factor of expansion/compression
>
> # Angles of points from center
> angles=seq(0, 2*pi*(20-1/points), length.out = points)+pi/2
>
> # Initial center
> df=data.frame(x=0, y=0)
>
> # Iterate over centers again and again
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+radius^(k-1)*cos(angles)*sin(4*angles),
+ y=df[i,"y"]+radius^(k-1)*sin(angles)*sin(2*angles)) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }
>
> # Obtain Voronoi regions
> df %>%
+ select(x,y) %>%
+ deldir(sort=TRUE) %>%
+ .$dirsgs -> data
>
> # Plot regions with geom_segmen
> data %>%
+ ggplot()
+ geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2), color="black") +
+ scale_x_continuous(expand=c(0,0))+
+ scale_y_continuous(expand=c(0,0))+
+ coord_fixed() +
+ theme(legend.position = "none",
+ panel.background = element_rect(fill="seagreen"),
+ panel.border = element_rect(colour = "black", fill=NA),
+ axis.ticks = element_blank(),
+ panel.grid = element_blank(),
+ axis.title = element_blank(),
+ axis.text = element_blank())->plot
>
> plot

data visulazition in R
> library(ggplot2)
> library(dplyr)
> library(deldir)
>
> # Parameters to change as you like
> iter=4 # Number of iterations (depth)
> points=7 # Number of points
> radius=3.5 # Factor of expansion/compression
>
> # Angles of points from center
> angles=seq(0, 2*pi*(10-1/points), length.out = points)+pi/2
>
> # Initial center
> df=data.frame(x=0, y=0)
>
> # Iterate over centers again and again
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+radius^(k-1)*cos(angles)*sin(4*angles),
+ y=df[i,"y"]+radius^(k-1)*sin(angles)*sin(2*angles)) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }
>
> # Obtain Voronoi regions
> df %>%
+ select(x,y) %>%
+ deldir(sort=TRUE) %>%
+ .$dirsgs -> data
>
> # Plot regions with geom_segmen
> data %>%
+ ggplot() + ggtitle("by Volkan OBAN using R - mandalas") +
+ geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2), color="black") +
+ scale_x_continuous(expand=c(0,0))+
+ scale_y_continuous(expand=c(0,0))+
+ coord_fixed() +
+ theme(legend.position = "none",
+ panel.background = element_rect(fill="plum2"),
+ panel.border = element_rect(colour = "black", fill=NA),
+ axis.ticks = element_blank(),
+ panel.grid = element_blank(),
+ axis.title = element_blank(),
+ axis.text = element_blank())->plot
>
> plot

Plot
> library(ggplot2)
> library(dplyr)
> library(deldir)
> # Parameters to change as you like
> iter=5 # Number of iterations (depth)
> points=7 # Number of points
> radius=4 # Factor of expansion/compression
> # Angles of points from center
> angles=seq(0, 2*pi*(2-1/points), length.out = points)+pi*pi/8
> # Initial center
> df=data.frame(x=0, y=0)
> # Iterate over centers again and again
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+radius^(k-1)*cos(angles)*pi,
+ y=df[i,"y"]+radius^(k-1)*sin(angles)*pi) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }
> # Obtain Voronoi regions
> df %>%
+ select(x,y) %>%
+ deldir(sort=TRUE) %>%
+ .$dirsgs -> data
> # Plot regions with geom_segmen
> data %>%
geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2), color="black") +
+ scale_x_continuous(expand=c(0,0))+
+ scale_y_continuous(expand=c(0,0))+
+ coord_fixed() +
+ theme(legend.position = "none",
+ panel.background = element_rect(fill="red"),
+ panel.border = element_rect(colour = "black", fill=NA),
+ axis.ticks = element_blank(),
+ panel.grid = element_blank(),
+ axis.title = element_blank(),
+ axis.text = element_blank())->plot
> plot
Plot
> library(ggplot2)
> library(dplyr)
> library(deldir)
> # Parameters to change as you like
> iter=5 # Number of iterations (depth)
> points=7 # Number of points
> radius=4 # Factor of expansion/compression
> # Angles of points from center
> angles=seq(0, 2*pi*(4-1/points), length.out = points)+pi/4
> # Initial center
> df=data.frame(x=0, y=0)
> # Iterate over centers again and again
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+radius^(k-1)*cos(angles)*pi*k-2,
+ y=df[i,"y"]+radius^(k-1)*sin(angles)*pi*k) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }
> # Obtain Voronoi regions
> df %>%
+ select(x,y) %>%
+ deldir(sort=TRUE) %>%
+ .$dirsgs -> data
> # Plot regions with geom_segmen
> data %>%
+ geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2), color="black") +
+ scale_x_continuous(expand=c(0,0))+
+ scale_y_continuous(expand=c(0,0))+
+ coord_fixed() +
+ theme(legend.position = "none",
+ panel.background = element_rect(fill="blue"),
+ panel.border = element_rect(colour = "black", fill=NA),
+ axis.ticks = element_blank(),
+ panel.grid = element_blank(),
+ axis.title = element_blank(),
+ axis.text = element_blank())->plot
> plot

data aRt with R -Mandalas
> library(ggplot2)
> library(dplyr)
> library(deldir)
> iter=5 # Number of iterations (depth)
> points=7 # Number of points
> radius=4 # F
> angles=seq(0, 2*pi*(1-1/points), length.out = points)+pi/2
> # Initial center
> df=data.frame(x=0, y=0)
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+radius^(k-1)*cos(angles)*4*k,
+ y=df[i,"y"]+radius^(k-1)*sin(angles)*2*k) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }
> # Obtain Voronoi regions
> df %>%
+ select(x,y) %>%
+ deldir(sort=TRUE) %>%
+ .$dirsgs -> data
> # Plot regions with geom_segmen
> data %>%
+ geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2), color="black") +
+ scale_x_continuous(expand=c(0,0))+
+ scale_y_continuous(expand=c(0,0))+
+ coord_fixed() +
+ theme(legend.position = "none",
+ panel.background = element_rect(fill="green"),
+ panel.border = element_rect(colour = "black", fill=NA),
+ axis.ticks = element_blank(),
+ panel.grid = element_blank(),
+ axis.title = element_blank(),
+ axis.text = element_blank())->plot
> plot

Plot
data.frame(x=df[i,"x"]+2*pi*radius^(k-1)*cos(angles)*sin(angles) ,
+ y=df[i,"y"]+2*pi*radius^(k-2)*sin(angles))

mandalas
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+2*pi*radius^(k-1)*cos(angles)+ sin(angles) ,
+ y=df[i,"y"]+2*pi*radius^(k-2)*sin(angles)) %>% rbind(temp) -> temp
+ }
+ df=temp

data-aRt
library(ggplot2)
> library(dplyr)
> library(deldir)
>
> iter=5 # Number of iterations (depth)
> points=7 # Number of points
> radius=3.6 # Factor of expansion/compression
>
> # Angles of points from center
> angles=seq(0, 2*pi*(1-1/points), length.out = points)+pi/2
> df=data.frame(x=0, y=0)
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+4*pi*radius^(k-3)*cos(angles)+ sin(angles) ,
+ y=df[i,"y"]+2*pi*radius^(k-2)*sin(angles)) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }
> df %>%
+ select(x,y) %>%
+ deldir(sort=TRUE) %>%
+ .$dirsgs -> data
> data %>%
+ ggplot() +
+ geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2), color="black") +
+ scale_x_continuous(expand=c(0,0))+
+ scale_y_continuous(expand=c(0,0))+
+ coord_fixed() +
+ theme(legend.position = "none",
+ panel.background = element_rect(fill="firebrick1"),
+ panel.border = element_rect(colour = "black", fill=NA),
+ axis.ticks = element_blank(),
+ panel.grid = element_blank(),
+ axis.title = element_blank(),
+ axis.text = element_blank())->plot
>
> plot

Plot
> library(ggplot2)
> library(dplyr)
> library(deldir)
>
> iter=3 # Number of iterations (depth)
> points=6 # Number of points
> radius=3.8 # Factor of expansion/compression
>
> angles=seq(0, 2*pi*(1-1/points), length.out = points)+pi/2
>
> df=data.frame(x=0, y=0)
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+4*pi*radius^(k-3)*cos(angles)+ sin(angles) ,
+ y=df[i,"y"]+2*pi*radius^(k-2)*sin(angles)) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }
>
s
> df %>%
+ select(x,y) %>%
+ deldir(sort=TRUE) %>%
+ .$dirsgs -> data
>
> data %>%
+ ggplot() + ggtitle((" Mandalas")) +
+ geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2), color="black") +
+ scale_x_continuous(expand=c(0,0))+
+ scale_y_continuous(expand=c(0,0))+
+ coord_fixed() +
+ theme(legend.position = "none",
+ panel.background = element_rect(fill="seagreen3"),
+ panel.border = element_rect(colour = "black", fill=NA),
+ axis.ticks = element_blank(),
+ panel.grid = element_blank(),
+ axis.title = element_blank(),
+ axis.text = element_blank())->plot
>
> plot
ref:https://github.com/aschinchon/mandalas/blob/master/mandala.R

mandalas
..... data.frame(x=df[i,"x"]+4*pi*radius^(k-1)*cos(angles) + sin(angles) ,
+ y=df[i,"y"]+2*pi*radius^(k-1)*sin(angles)) %>% rbind(temp) -> temp..............

mandalas
> library(ggplot2)
> library(dplyr)
> library(deldir)
>
> # Parameters to change as you like
> iter=3 # Number of iterations (depth)
> points=6 # Number of points
> radius=3.8 # Factor of expansion/compression
>
> # Angles of points from center
> angles=seq(0, 2*pi*(1-1/points), length.out = points)+pi/2
>
> # Initial center
> df=data.frame(x=0, y=0)
>
> # Iterate over centers again and again
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+4*radius^(k-1)*cos(angles),
+ y=df[i,"y"]+2*radius^(k-1)*sin(angles)) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }
>
> # Obtain Voronoi regions
> df %>%
+ select(x,y) %>%
+ deldir(sort=TRUE) %>%
+ .$dirsgs -> data
>
> # Plot regions with geom_segmen
> data %>%
+ ggplot() + ggtitle((" Mandalas")) +
+ geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2), color="black") +
+ scale_x_continuous(expand=c(0,0))+
+ scale_y_continuous(expand=c(0,0))+
+ coord_fixed() +
+ theme(legend.position = "none",
+ panel.background = element_rect(fill="violetred4"),
+ panel.border = element_rect(colour = "black", fill=NA),
+ axis.ticks = element_blank(),
+ panel.grid = element_blank(),
+ axis.title = element_blank(),
+ axis.text = element_blank())->plot
>
> plot

mandalas
> library(ggplot2)
> library(dplyr)
> library(deldir)
>
> # Parameters to change as you like
> iter=3 # Number of iterations (depth)
> points=6 # Number of points
> radius=3.8 # Factor of expansion/compression
>
> # Angles of points from center
> angles=seq(0, 2*pi*(1-1/points), length.out = points)+pi/2
>
> # Initial center
> df=data.frame(x=0, y=0)
>
> # Iterate over centers again and again
> for (k in 1:iter)
+ {
+ temp=data.frame()
+ for (i in 1:nrow(df))
+ {
+ data.frame(x=df[i,"x"]+radius^(k-1)*cos(angles),
+ y=df[i,"y"]+2*radius^(k-1)*sin(angles)) %>% rbind(temp) -> temp
+ }
+ df=temp
+ }
>
> # Obtain Voronoi regions
> df %>%
+ select(x,y) %>%
+ deldir(sort=TRUE) %>%
+ .$dirsgs -> data
>
> # Plot regions with geom_segmen
> data %>%
+ ggplot() + ggtitle((" by Volkan OBAN using R - Mandalas")) +
+ geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2), color="black") +
+ scale_x_continuous(expand=c(0,0))+
+ scale_y_continuous(expand=c(0,0))+
+ coord_fixed() +
+ theme(legend.position = "none",
+ panel.background = element_rect(fill="royalblue1"),
+ panel.border = element_rect(colour = "black", fill=NA),
+ axis.ticks = element_blank(),
+ panel.grid = element_blank(),
+ axis.title = element_blank(),
+ axis.text = element_blank())->plot
>
> plot

mandalas
library(ggplot2)
library(dplyr)
library(deldir)
# Parameters to change as you like
iter=5 # Number of iterations (depth)
points=7 # Number of points
radius=3.8 # Factor of expansion/compression
# Angles of points from center
angles=seq(0, 2*pi*(1-1/points), length.out = points)+pi/2
# Initial center
df=data.frame(x=0, y=0)
# Iterate over centers again and again
for (k in 1:iter)
{
temp=data.frame()
for (i in 1:nrow(df))
{
data.frame(x=df[i,"x"]+radius^(k-1)*sin(angles)*cos(angles),
y=df[i,"y"]+radius^(k-1)*sin(angles)) %>% rbind(temp) -> temp
}
df=temp
}
# Obtain Voronoi regions
df %>%
select(x,y) %>%
deldir(sort=TRUE) %>%
.$dirsgs -> data
# Plot regions with geom_segmen
data %>%
ggplot() + ggtitle("Mandalas") +
geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2), color="black") +
scale_x_continuous(expand=c(0,0))+
scale_y_continuous(expand=c(0,0))+
coord_fixed() +
theme(legend.position = "none",
panel.background = element_rect(fill="lightsteelblue"),
panel.border = element_rect(colour = "black", fill=NA),
axis.ticks = element_blank(),
panel.grid = element_blank(),
axis.title = element_blank(),
axis.text = element_blank())->plot
plot

ggplot2 and ggthemr
> ggthemr('chalk')
> library(ggthemes)
> g <- ggplot(mpg, aes(class, cty))
> g + geom_boxplot(aes(fill=factor(cyl))) +
+ theme(axis.text.x = element_text(angle=65, vjust=0.6)) +
+ labs(title=" - ggtherm and ggplot2",
+ subtitle="City Mileage grouped by Class of vehicle",
+ caption="Source: mpg",
+ x="Class of Vehicle",
+ y="City Mileage")

ggplot2 and ggthemr
ggthemr('earth')
> library(ggthemes)
> g <- ggplot(mpg, aes(class, cty))
> g + geom_boxplot(aes(fill=factor(cyl))) +
+ theme(axis.text.x = element_text(angle=65, vjust=0.6)) +
+ labs(title=" ggtherm and ggplot2",
+ subtitle="City Mileage grouped by Class of vehicle",
+ caption="Source: mpg",
+ x="Class of Vehicle",
+ y="City Mileage")
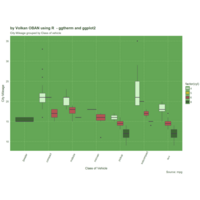
ggthemr
ggthemr('grass')
> library(ggthemes)
> g <- ggplot(mpg, aes(class, cty))
> g + geom_boxplot(aes(fill=factor(cyl))) +
+ theme(axis.text.x = element_text(angle=65, vjust=0.6)) +
+ labs(title=" ggtherm and ggplot2",
+ subtitle="City Mileage grouped by Class of vehicle",
+ caption="Source: mpg",
+ x="Class of Vehicle",
+ y="City Mileage")

Plot
library(ggthemes)
ggthemr('sea)
> g <- ggplot(mpg, aes(class, cty))
> g + geom_boxplot(aes(fill=factor(cyl))) +
+ theme(axis.text.x = element_text(angle=65, vjust=0.6)) +
+ labs(title="ggtherm and ggplot2",
+ subtitle="City Mileage grouped by Class of vehicle",
+ caption="Source: mpg",
+ x="Class of Vehicle",
+ y="City Mileage")
hexbin
> x <- rnorm(10000)
> y <- rnorm(10000)
> bin <- hexbin(x,y)
> ## Plot method for hexbin !
> ## ---- ------ --------
> plot(bin)
> # nested lattice
> plot(bin,, style= "nested.lattice")
hexbin
> set.seed(153)
> x <- rnorm(100000)
> y <- rnorm(100000)
> bin <- hexbin(x,y)
> smbin <- smooth.hexbin(bin)
> erodebin <- erode.hexbin(smbin, cdfcut=.4)
> plot(erodebin,main = "")

yarrr
Show in New WindowClear OutputExpand/Collapse Output
shiny.tag
shiny.tag
shiny.tag
shiny.tag
shiny.tag
shiny.tag
shiny.tag
shiny.tag
Show in New WindowClear OutputExpand/Collapse Output
Error: unexpected symbol in:
"
print(p)Show"
Modify Chunk OptionsRun All Chunks AboveRun Current ChunkModify Chunk OptionsRun All Chunks AboveRun Current ChunkModify Chunk OptionsRun All Chunks AboveRun Current Chunk
Console~/
> pirateplot(formula = budget ~ creative.type,
+ data = subset(movies, budget > 0 &
+ creative.type %in% c("Multiple Creative Types", "Factual") == FALSE),
+ point.o = .02,
+ xlab = "",
+ main = " Data visualization with R - yarrr ",
+ gl.col = "gray",
+ pal = "black")
>
> mtext("Movie budgets (in millions) by rating -- pirateplot",
+ side = 3,
+ font = 3)
>
> mtext("*movies tend to have the highest budgets\n...by far!",
+ side = 1, adj = 1, line = 3,
+ cex = .8, font = 3)
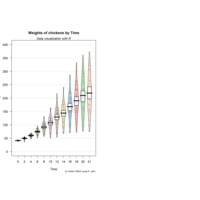
yarrr
pirateplot(formula = weight ~ Time,
data = ChickWeight,
main = "Weights of chickens by Time",
pal = "xmen",
gl.col = "gray")
mtext(text = "Using the xmen palette!",
side = 3,
font = 3)
mtext(text = "*The mean and variance of chicken\nweights tend to increase over time.",
side = 1,
adj = 1,
line = 3.5,
font = 3,
cex = .7)

swatches
ref: https://www.r-bloggers.com/new-package-swatches-is-now-on-cran/
library(swatches)
library(hrbrthemes)
library(tidyverse)
download.file("https://www.pantone.com/images/pages/21348/adobe-ase/Pantone-COY18-Palette-ASE-files.zip", "ultra_violet.zip")
unique(dirname((unzip("ultra_violet.zip"))))
## [1] "./Pantone COY18 Palette ASE files"
## [2] "./__MACOSX/Pantone COY18 Palette ASE files"
dir("./Pantone COY18 Palette ASE files")
par(mfrow=c(8,1))
dir("./Pantone COY18 Palette ASE files", full.names=TRUE) %>%
walk(~{
pal_name <- gsub("(^[[:alnum:]]+-|\\.ase$)", "", basename(.x))
show_palette(read_palette(.x))
title(pal_name)
})
par(mfrow=c(1,1))
(intrigue <- read_palette("./Pantone COY18 Palette ASE files/PantoneCOY18-Intrigue.ase"))
(intrigue <- read_palette("./Pantone COY18 Palette ASE files/PantoneCOY18-Intrigue.ase", use_names=FALSE))
ggplot(economics_long, aes(date, value)) +
geom_area(aes(fill=variable)) +
scale_y_comma() +
scale_fill_manual(values=intrigue) +
facet_wrap(~variable, scales = "free", nrow = 2, strip.position = "bottom") +
theme_ipsum_rc(grid="XY", strip_text_face="bold") +
theme(strip.placement = "outside") +
theme(legend.position=c(0.85, 0.2))

PDN-Personalized Disease Network
#Select a subset of data for toy example
comorbidity_data = comorbidity_data[c(1:10),]
survival_data = survival_data[c(1:10),]
# Find date cuts
k1 = datecut(comorbidity_data,survival_data[,1],survival_data[,2])
# Build networks
a = buildnetworks(comorbidity_data,k1)
# Graph individual patients
datark = t(apply(comorbidity_data,1,rank))
dak = sort(datark[1,])
# draw PDN for the first patient
draw.PDN.circle(a[1,],dak)
# draw PDN for the whole comorbidity data set
par(mfrow=c(2,5))
for(i in 1 : nrow(a)){
dak = apply(datark,2,sort)
draw.PDN.circle(a[i,],dak[i,])
title(main=paste("Patient",i))
}
Plot
library(ggplot2)
library(ggthemes)
> theme_set(theme_bw())
> g <- ggplot(mpg, aes(manufacturer, cty))
> g + geom_boxplot() +
+ geom_dotplot(binaxis='y',
+ stackdir='center',
+ dotsize = .5,
+ fill="yellow") +
+ theme(axis.text.x = element_text(angle=65, vjust=0.6)) +
+ labs(title=" ",
+ caption="Data visualization with R",
+ x="Class of Vehicle",
+ y="City Mileage") +theme_hc(bgcolor = "darkunica") +
+ scale_fill_hc("darkunica"
sunflowerplot
sunflowerplot(rnorm(1000), rnorm(1000), number = rpois(n = 1000, lambda = 2),rotate = TRUE, col = "purple")

animation
> library(ggplot2)
> library(dplyr)
> library(tidyr)
> library(purrr)
> library(animation)
> library(gganimate)
>
> ## Function to evaluate Beta pdf for a vector of values ##
> calc_beta = function(alpha, beta){
+ x = seq(0.01, 0.99, 0.01)
+ densityf = dbeta(x, shape = alpha, shape2 = beta)
+ return(data_frame(x, densityf))
+ }
>
> ## Create data frame with evaluation of Beta pdf for different combinations of alpha and beta ##
> alpha = c(0.1, 0.5, 1:5, 10)
> beta = c(0.5, 1, 2, 5)
>
> ## Create data frame ##
> # Couldn't get the pipe operator to properly show up in WordPress :-(
> df = expand.grid(alpha = alpha, beta = beta)
> df = group_by(df, alpha, beta)
> df = unnest(mutate(df, plotdata = map2(alpha, beta, calc_beta)))
>
> ## Create plot ##
> p = ggplot(df, aes(x = x, y = densityf, colour = factor(alpha), group = factor(alpha))) + ggtitle("by Volkan OBAN using R ")+
+ geom_path(aes(frame = alpha, cumulative = TRUE), size = 0.5) +
+ facet_wrap(~beta,
+ labeller = label_bquote(cols = beta == .(beta))) +
+ ylim(c(0, 6)) +
+ labs(y = expression(paste("f(x; ", alpha, ", ", beta, ")")),
+ title = "Changing parameters in Beta density function") +
+ scale_colour_discrete(name = expression(alpha)) +
+ theme(plot.title = element_text(hjust = 0.5))
Warning: Ignoring unknown aesthetics: frame, cumulative
>
> ani.options(interval = 0.8)
> gganimate(p, title_frame = FALSE, width = 4, height = 4)
reference: http://www.masterdataanalysis.com/r/creating-animations-ggplot2-plots/

tweenr
> library(ggplot2)
> library(gganimate)
> library(ggforce)
> library(tweenr)
>
> # Making up data
> d <- data.frame(x = rnorm(20), y = rnorm(20), time = sample(100, 20), alpha = 0,
+ size = 1, ease = 'elastic-out', id = 1:20,
+ stringsAsFactors = FALSE)
> d2 <- d
> d2$time <- d$time + 10
> d2$alpha <- 1
> d2$size <- 3
> d2$ease <- 'linear'
> d3 <- d2
> d3$time <- d2$time + sample(50:100, 20)
> d3$size = 10
> d3$ease <- 'bounce-out'
> d4 <- d3
> d4$y <- min(d$y) - 0.5
> d4$size <- 2
> d4$time <- d3$time + 10
> d5 <- d4
> d5$time <- max(d5$time)
> df <- rbind(d, d2, d3, d4, d5)
>
> # Using tweenr
> dt <- tween_elements(df, 'time', 'id', 'ease', nframes = 500)
>
> # Animate with gganimate
> p <- ggplot(data = dt) +
+ geom_point(aes(x=x, y=y, size=size, alpha=alpha, frame = .frame)) +
+ scale_size(range = c(0.1, 20), guide = 'none') +
+ scale_alpha(range = c(0, 1), guide = 'none') +
+ ggforce::theme_no_axes()
Warning: Ignoring unknown aesthetics: frame
> animation::ani.options(interval = 1/24)
> gganimate(p, 'dropping balls.gif', title_frame = F)

tweenr
library(ggplot2)
> library(gganimate)
> library(ggforce)
> library(tweenr)
>
> # Making up data
> t <- data.frame(x=0, y=0, colour = 'forestgreen', size=1, alpha = 1,
+ stringsAsFactors = FALSE)
> t <- t[rep(1, 12),]
> t$alpha[2:12] <- 0
> t2 <- t
> t2$y <- 1
> t2$colour <- 'firebrick'
> t3 <- t2
> t3$x <- 1
> t3$colour <- 'steelblue'
> t4 <- t3
> t4$y <- 0
> t4$colour <- 'goldenrod'
> t5 <- t4
> c <- ggforce::radial_trans(c(1,1), c(1, 12))$transform(rep(1, 12), 1:12)
> t5$x <- (c$x + 1) / 2
> t5$y <- (c$y + 1) / 2
> t5$alpha <- 1
> t5$size <- 0.5
> t6 <- t5
> t6 <- rbind(t5[12,], t5[1:11, ])
> t6$colour <- 'firebrick'
> t7 <- rbind(t6[12,], t6[1:11, ])
> t7$colour <- 'steelblue'
> t8 <- t7
> t8$x <- 0.5
> t8$y <- 0.5
> t8$size <- 2
> t9 <- t
> ts <- list(t, t2, t3, t4, t5, t6, t7, t8, t9)
>
> tweenlogo <- data.frame(x=0.5, y=0.5, label = 'tweenr', stringsAsFactors = F)
> tweenlogo <- tweenlogo[rep(1, 60),]
> tweenlogo$.frame <- 316:375
>
> # Using tweenr
> tf <- tween_states(ts, tweenlength = 2, statelength = 1,
+ ease = c('cubic-in-out', 'elastic-out', 'bounce-out',
+ 'cubic-out', 'sine-in-out', 'sine-in-out',
+ 'circular-in', 'back-out'),
+ nframes = 375)
>
> # Animate with gganimate
> p <- ggplot(data=tf, aes(x=x, y=y)) +
+ geom_text(aes(label = label, frame = .frame), data=tweenlogo, size = 13) +
+ geom_point(aes(frame = .frame, size=size, alpha = alpha, colour = colour)) +
+ scale_colour_identity() +
+ scale_alpha(range = c(0, 1), guide = 'none') +
+ scale_size(range = c(4, 60), guide = 'none') +
+ expand_limits(x=c(-0.36, 1.36), y=c(-0.36, 1.36)) +
+ theme_bw()
Warning: Ignoring unknown aesthetics: frame
Warning: Ignoring unknown aesthetics: frame
> animation::ani.options(interval = 1/15)
> gganimate(p, "dancing ball.gif", title_frame = F, ani.width = 400,
+ ani.height = 400)
D3partitionR
d3 = D3partitionR() %>%
add_data(data_plot,count = 'N',tooltip=c('name','Location'),steps=c('Sex','Embarked','Pclass','Survived')) %>%
add_nodes_data(list('Embarked S'=list('Location'='<a href="https://fr.wikipedia.org/wiki/Southampton">Southampton</a>'),
'Embarked C'=list('Location'='<a href="https://fr.wikipedia.org/wiki/Cherbourg-Octeville">Cherbourg</a>'),
'Embarked Q'=list('Location'='<a href="https://fr.wikipedia.org/wiki/Cobh">Queenstown</a>')
)
)
d3 %>%
set_legend_parameters(zoom_subset = TRUE) %>%
set_chart_type('circle_treemap') %>%
set_tooltip_parameters(visible=TRUE, style='background-color:lightblue;',builder='basic') %>%
plot()

plotly
library(ggplot2)
> data.diamonds=ggplot2::diamonds
> library(plotly)
> gg=ggplot(data.diamonds,aes(x=carat,y=price,color=color))+geom_point(alpha=0.3)
> ggplotly(gg)
ggiraph
dataset = data.frame( x1 = c(1, 5, 1, 3, 0),
+ x2 = c(2, 4, 0, 4, 5),
+ y1 = c( 1, 8, 0, 1, 3),
+ y2 = c( 2, 2, 5, 3, 4),
+ t = c( 'O', 'O', 'O', 'V', 'V'),
+ r = c( 1, 2, 3, 4, 5),
+ tooltip = c("ID 1", "ID 2", "ID 3", "ID 4", "ID 5"),
+ uid = c("ID 1", "ID 2", "ID 3", "ID 4", "ID 5"),
+ oc = rep("alert(this.getAttribute(\"data-id\"))", 5)
+ )
>
> gg_rect = ggplot() +
+ scale_x_continuous(name="x ") +
+ scale_y_continuous(name="y") +
+ geom_rect_interactive(data=dataset,
+ mapping = aes(xmin = x1, xmax = x2,
+ ymin = y1, ymax = y2, fill = t,
+ tooltip = tooltip, onclick = oc, data_id = uid ),
+ color="purple", alpha=0.6) +
+ geom_text(data=dataset,
+ aes(x = x1 + ( x2 - x1 ) / 2, y = y1 + ( y2 - y1 ) / 2,
+ label = r ),
+ size = 4 )
>
>
> ggiraph(code = {print(gg_rect)})

ggiraph
p <- ggplot(mpg, aes(x = drv, y = hwy, tooltip = class, fill = class)) +
+ geom_boxplot_interactive(outlier.colour = "blue") +
guides(fill = "none") + theme_minimal()
> ggiraph(code = print(p))

heatmap
ggplot(train, aes(Outlet_Identifier, Item_Type))+
+ geom_raster(aes(fill = Item_MRP))+
+ labs(title =" Heat Map", x = "Outlet Identifier", y = "Item Type")+
+ scale_fill_continuous(name = "Item MRP")

ggplot2
> ggplot(train, aes(Outlet_Identifier, Item_Outlet_Sales)) + geom_boxplot(fill = "yellow")+
+ scale_y_continuous("Item Outlet Sales", breaks= seq(0,15000, by=500))+
+ labs(title = "", x = "Outlet Identifier")
data<-https://docs.google.com/spreadsheets/d/1PR5StHxg2jlMCb4IUilGSEwhylXn-3q3EJucSaVolCU/edit#gid=0

ggplot2 and ggthemes
> yearly_weight <- surveys_complete %>%
+ group_by(year, species_id, sex) %>%
+ summarise(avg_weight = mean(weight, na.rm = TRUE))
> ggplot(yearly_weight, aes(x=year, y=avg_weight, color = sex, group = sex)) +
+ geom_line() +
+ facet_wrap(~ species_id) + theme_solarized() +
+ scale_colour_solarized("blue")
ggplot2 and ggthemes
> ggplot(surveys_complete, aes(x = species_id, y = hindfoot_length)) +
+ geom_boxplot(alpha = 0) +
+ geom_jitter(alpha = 0.3, color = "yellow")+ theme_solarized_2(light = FALSE) +
+ scale_colour_solarized("blue")
Plot3d
ref
https://cran.r-project.org/web/packages/plot3D/vignettes/plot3D.pdf

Plot3d
ref: https://cran.r-project.org/web/packages/plot3D/vignettes/plot3D.pdf

plot3D
> x <- (3 + cos(2*v)*sin(2*u) - sin(3*v)*sin(2*u))*cos(v)
> y <- (3 + cos(v)*sin(u) - sin(v)*sin(3*u))*sin(v);z <- sin(v)*sin(2*u) + cos(v)*sin(u)
> surf3D(x, y, z,,colvar = z, colkey = FALSE, facets = FALSE)

plot3D
> x <- (3 + cos(v/2)*sin(u) - sin(v/2)*sin(2*u))*cos(v)
> y <- (3 + cos(v/2)*sin(u) - sin(v/2)*sin(2*u))*sin(v)
> z <- sin(2*v)*sin(u) + cos(2*v)*sin(2*u)
> surf3D(x, y, z, colvar = z, colkey = FALSE, facets = FALSE)

plot3D
> M <- mesh(seq(0, 6*pi, length.out = 80), seq(pi/3, pi, length.out = 80))
> u <- M$x ; v <- M$y
> x <- u/2 * cos(2*v)
> y <- u/2 * sin(v) * sin(2*u)
> z <- u/2 * sin(2*v)
> surf3D(x, y, z, colvar = z,colkey = FALSE, box = FALSE)

ggplot2
library(tidyverse)
library(viridis)
library(OECD)
# search by keyword
search_dataset("unemployment") %>% View
# download the selected dataset
df_oecd <- get_dataset("AVD_DUR")
# turn variable names to lowercase
names(df_oecd) <- names(df_oecd) %>% tolower()
df_oecd %>%
filter(country %in% c("EU16", "EU28", "USA"), sex == "MEN", ! age == "1524") %>%
ggplot(aes(obstime, age, fill = obsvalue))+
geom_tile()+
scale_fill_viridis("Months", option = "B")+
scale_x_discrete(breaks = seq(1970, 2015, 5) %>% paste)+
facet_wrap(~ country, ncol = 1)+
labs(x = NULL, y = "Age groups",
title = "Average duration of unemployment in months, males")+
theme_minimal()
Clifford Attractors
> library("compiler")
>
> mapxy <- function(x, y, xmin, xmax, ymin=xmin, ymax=xmax) {
+ sx <- (width - 1) / (xmax - xmin)
+ sy <- (height - 1) / (ymax - ymin)
+ row0 <- round( sx * (x - xmin) )
+ col0 <- round( sy * (y - ymin) )
+ col0 * height + row0 + 1
+ }
>
> dejong <- function(x, y) {
+ nidxs <- length(mat)
+ counts <- integer(length=nidxs)
+ for (i in 1:npoints) {
+ xt <- sin(a * y) - cos(b * x)
+ y <- sin(c * x) - cos(d * y)
+ x <- xt
+ idxs <- mapxy(x, y, -2, 2)
+ counts <- counts + tabulate(idxs, nbins=nidxs)
+ }
+ mat <<- mat + counts
+ }
>
> clifford <- function(x, y) {
+ ac <- abs(c)+1
+ ad <- abs(d)+1
+ nidxs <- length(mat)
+ counts <- integer(length=nidxs)
+ for (i in 1:npoints) {
+ xt <- sin(a * y) + c * cos(a * x)
+ y <- sin(b * x) + d * cos(b * y)
+ x <- xt
+ idxs <- mapxy(x, y, -ac, ac, -ad, ad)
+ counts <- counts + tabulate(idxs, nbins=nidxs)
+ }
+ mat <<- mat + counts
+ }
>
> #color vector
> cvec <- grey(seq(0, 1, length=10))
> #can also try other colours, see help(rainbow)
> #cvec <- heat.colors(10)
>
> #we end up with npoints * n points
> npoints <- 8
> n <- 100000
> width <- 600
> height <- 600
>
> #make some random points
> rsamp <- matrix(runif(n * 2, min=-2, max=2), nr=n)
>
> #compile the functions
> setCompilerOptions(suppressAll=TRUE)
> mapxy <- cmpfun(mapxy)
> dejong <- cmpfun(dejong)
> clifford <- cmpfun(clifford)
>
> #dejong
> a <- 1.4
> b <- -2.3
> c <- 2.4
> d <- -2.1
>
> mat <- matrix(0, nr=height, nc=width)
> dejong(rsamp[,1], rsamp[,2])
>
> #this applies some smoothing of low valued points, from A.N. Spiess
> #QUANT <- quantile(mat, 0.5)
> #mat[mat <= QUANT] <- 0
>
> dens <- log(mat + 1)/log(max(mat))
> par(mar=c(0, 0, 0, 0))
> image(t(dens), col=cvec, useRaster=T, xaxt='n', yaxt='n')
>
> #clifford
> a <- -1.4
> b <- 1.6
> c <- 1.0
> d <- 0.7
>
> mat <- matrix(0, nr=height, nc=width)
> #QUANT <- quantile(mat, 0.5)
> #mat[mat <= QUANT] <- 0
> clifford(rsamp[,1], rsamp[,2])
>
> dens <- log(mat + 1)/log(max(mat))
> par(mar=c(0, 0, 0, 0))
> image(t(dens), col=cvec, useRaster=T, xaxt='n', yaxt='n')
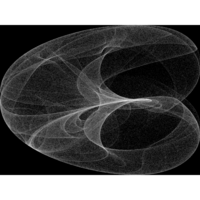
Clifford Attractors
library("compiler")
>
> mapxy <- function(x, y, xmin, xmax, ymin=xmin, ymax=xmax) {
+ sx <- (width - 1) / (xmax - xmin)
+ sy <- (height - 1) / (ymax - ymin)
+ row0 <- round( sx * (x - xmin) )
+ col0 <- round( sy * (y - ymin) )
+ col0 * height + row0 + 1
+ }
>
> dejong <- function(x, y) {
+ nidxs <- length(mat)
+ counts <- integer(length=nidxs)
+ for (i in 1:npoints) {
+ xt <- sin(a * y) - cos(b * x)
+ y <- sin(c * x) - cos(d * y)
+ x <- xt
+ idxs <- mapxy(x, y, -2, 2)
+ counts <- counts + tabulate(idxs, nbins=nidxs)
+ }
+ mat <<- mat + counts
+ }
>
> clifford <- function(x, y) {
+ ac <- abs(c)+1
+ ad <- abs(d)+1
+ nidxs <- length(mat)
+ counts <- integer(length=nidxs)
+ for (i in 1:npoints) {
+ xt <- sin(a * y) + c * cos(a * x)
+ y <- sin(b * x) + d * cos(b * y)
+ x <- xt
+ idxs <- mapxy(x, y, -ac, ac, -ad, ad)
+ counts <- counts + tabulate(idxs, nbins=nidxs)
+ }
+ mat <<- mat + counts
+ }
>
> #color vector
> cvec <- grey(seq(0, 1, length=10))
> #can also try other colours, see help(rainbow)
> #cvec <- heat.colors(10)
>
> #we end up with npoints * n points
> npoints <- 8
> n <- 100000
> width <- 600
> height <- 600
>
> #make some random points
> rsamp <- matrix(runif(n * 2, min=-2, max=2), nr=n)
>
> #compile the functions
> setCompilerOptions(suppressAll=TRUE)
> mapxy <- cmpfun(mapxy)
> dejong <- cmpfun(dejong)
> clifford <- cmpfun(clifford)
>
> #dejong
> a <- 1.4
> b <- -2.3
> c <- 2.4
> d <- -2.1
>
> mat <- matrix(0, nr=height, nc=width)
> dejong(rsamp[,1], rsamp[,2])
>
> #this applies some smoothing of low valued points, from A.N. Spiess
> #QUANT <- quantile(mat, 0.5)
> #mat[mat <= QUANT] <- 0
>
> dens <- log(mat + 1)/log(max(mat))
> par(mar=c(0, 0, 0, 0))
> image(t(dens), col=cvec, useRaster=T, xaxt='n', yaxt='n')
>
> #clifford
> a <- -1.4
> b <- 1.6
> c <- 1.0
> d <- 0.7
>
> mat <- matrix(0, nr=height, nc=width)
> #QUANT <- quantile(mat, 0.5)
> #mat[mat <= QUANT] <- 0
> clifford(rsamp[,1], rsamp[,2])
>
> dens <- log(mat + 1)/log(max(mat))
> par(mar=c(0, 0, 0, 0))
> image(t(dens), col=cvec, useRaster=T, xaxt='n', yaxt='n')
ref:https://github.com/petewerner/misc/blob/master/attractor.R

mvmesh
plot( SolidRectangle( a=c(1,3), b=c(2,7),
+ breaks=list( seq(1,3,by=0.25), seq(2,7,by=1) ) ), show.labels=TRUE

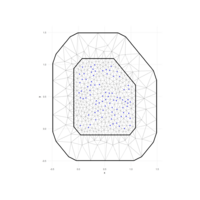
RTriangle
> p <- pslg(P=rbind(c(0, 0), c(0, 1), c(0.5, 0.5), c(1, 1), c(1, 0)),
+ S=rbind(c(1, 2), c(2, 3), c(3, 4), c(4, 5), c(5, 1)))
> ## Plot it
> plot(p)
> ## Triangulate it
> tp <- triangulate(p)
>
> ## Triangulate it subject to minimum area constraint
> tp <- triangulate(p, a=0.01)
> plot(tp)

plotmo
if (require(gbm)) {
n <- 100 # toy model for quick demo
x1 <- 3 * runif(n)
x2 <- 3 * runif(n)
x3 <- sample(1:4, n, replace=TRUE)
y <- x1 + x2 + x3 + rnorm(n, 0, .3)
data <- data.frame(y=y, x1=x1, x2=x2, x3=x3)
mod <- gbm(y~., data=data, distribution="gaussian",
n.trees=300, shrinkage=.1, interaction.depth=3,
train.fraction=.8, verbose=FALSE)
plot_gbm(mod)
# plotres(mod) # plot residuals
# plotmo(mod) # plot regression surfaces
}

rpart.plot
tree1 <- rpart(survived~., data=ptitanic)
par(mfrow=c(4,3))
for(iframe in 1:nrow(tree1$frame)) {
cols <- ifelse(1:nrow(tree1$frame) <= iframe, "black", "gray")
prp(tree1, col=cols, branch.col=cols, split.col=cols)
}

rpart.plot
data(ptitanic)
tree <- rpart(age ~ ., data=ptitanic)
rpart.plot(tree, type=4, extra=0, branch.lty=3, box.palette="RdYlGn")

brownian motion
>t <- 0:100 # time
> sig2 <- 0.01
> nsim <- 1000
> ## we'll simulate the steps from a uniform distribution with limits set to
> ## have the same variance (0.01) as before
> X <- matrix(runif(n = nsim * (length(t) - 1), min = -sqrt(3 * sig2), max = sqrt(3 * sig2)), nsim, length(t) - 1)
> X <- cbind(rep(0, nsim), t(apply(X, 1, cumsum)))
> plot(t, X[1, ],xlab = "time", ylab = "y",col="red", ylim = c(-2, 2), type = "l")
> apply(X[2:nsim, ], 1, function(x, t) lines(t, x), t = t)

persp
> x <- seq(-10, 10, length= 30)
> y <- x
> f <- function(x, y) { r <- sqrt(5*x^(-2)+exp(y^2)); 10 * sin(sin(r))/r }
> z <- outer(x, y, f)
> z[is.na(z)] <- 1
> op <- par(bg = "purple")
> persp(x, y, z, theta =70 , phi = 40, expand = 0.5, col = "yellow")

persp
op <- par(bg = "black")
> persp(x, y, z, theta =70 , phi = 40, expand = 0.5, col = "red")
persp-- Perspective Plots
> x <- seq(-10, 10, length= 30)
> y <- x
> f <- function(x, y) { r <- sqrt(5*x^(-2)+exp(y^2)); 10 * sin(sin(r))/r }
> z <- outer(x, y, f)
> z[is.na(z)] <- 1
> op <- par(bg = "gray")
> persp(x, y, z, theta =60 , phi = 30, expand = 0.5, col = "red")
persp-- Perspective Plots
> x <- seq(-10, 10, length= 30)
> y <- x
> f <- function(x, y) { r <- sqrt(x^(-2)+y^2); 10 * sin(cos(r))/r }
> z <- outer(x, y, f)
> z[is.na(z)] <- 1
> op <- par(bg = "white")
persp(x, y, z , theta =60 , phi = 30, expand = 0.5, col = "purple")
persp-- Perspective Plots
x <- seq(-10, 10, length= 30)
y <- x
> f <- function(x, y) { r <- sqrt(x^(-2)+y^2); 10 * sin(cos(r))/r }
> z <- outer(x, y, f)
> z[is.na(z)] <- 1
> op <- par(bg = "white")
> persp(x, y, z, theta = 30, phi = 30, expand = 0.5, col = "red")

lattice - wireframe
> g <- function(x, y) {(1 + y * 2) ^ (-x^2 / y^3) * (1 + y * 1) ^ (x / y)}
>
> require(lattice)
> myRange = seq(0.01, 2, len = 30)
> grid <- expand.grid(x = myRange , y = myRange)
> grid$z <- g(grid$x, grid$y)
> print(wireframe(z ~ x * y",col="purple", grid))

lattice - wireframe
> g <- function(x, y) {(1 + y * 2) ^ (-x / y) * (1 + y * 1) ^ (x / y)}
> require(lattice)
> myRange = seq(0.01, 2, len = 80)
> grid <- expand.grid(x = myRange , y = myRange)
> grid$z <- g(grid$x, grid$y)
> print(wireframe(z ~ x * y,col="purple", grid))
Plot
> a <- 2
> b <- 3
> theta <- seq(0,10*pi,0.01)
> r <- a + b*theta
> data<- data.frame(x=r*cos(theta), y=r*sin(theta)) # Cartesian coords
> library(ggplot2)
> ggplot(data, aes(x,y)) + geom_point(col='green')

Plot
> golden.ratio = (sqrt(5) + 1)/2
> fibonacci.angle=360/(golden.ratio^2)
> c=1
> num_points=630
> x=rep(0,num_points)
> y=rep(0,num_points)
>
> for (n in 1:num_points) {
+ r=c*sqrt(n)
+ theta=fibonacci.angle*(n)
+ x[n]=r*cos(theta)
+ y[n]=r*sin(theta)
+ }
> plot(x,y,axes=FALSE,ann=FALSE,pch=19,cex=1)

ade4
> data (euro123)
> par(mfrow = c(2,2))
> triangle.plot(euro123$in78, clab = 0, cpoi = 2, addmean = TRUE,
+ show = FALSE)
> triangle.plot(euro123$in86, label = row.names(euro123$in78), clab = 0.8)
> triangle.biplot(euro123$in78, euro123$in86)
> triangle.plot(rbind.data.frame(euro123$in78, euro123$in86), clab = 1, addaxes = TRUE, sub = "Principal axis", csub = 2, possub = "topright")
> par(mfrow = c(1,1))

hexbin-hexplom
data(NHANES)
hexplom(NHANES[,9:13], xbins = 20,colramp = BTY, upper.panel = panel.hexboxplot)
lattice - wireframe
> x <- seq(-pi, pi, len = 20)
> y <- seq(-pi, pi, len = 20)
> g <- expand.grid(x = x, y = y)
> g$z <- cos(sqrt(g$x^2 + g$y^2))
> wireframe(z ~ x * y, g, drape = TRUE,
+ aspect = c(3,1), colorkey = TRUE

Plot persp
> x <- y <- seq(-5, 5, length= 20)
> f <- function(x,y){ z <- x^4 + y^3 -3 }
> z <- outer(x,y,f)
> persp(x, y, z,theta = 60, phi = 45, expand = 0.5, col = "purple")
>
Plot
> x <- y <- seq(-5, 5, length= 20)
> f <- function(x,y){ z <- x*2 + y^3 -3 }
> z <- outer(x,y,f)
> persp(x, y, z,theta = 60, phi = 45, expand = 0.5, col = "red")
Plot- persp
> x <- y <- seq(-5, 5, length= 20)
> f <- function(x,y){ z <- x*2 + y -3 }
> z <- outer(x,y,f)
> persp(x, y, z, theta = 30, phi = 30, expand = 0.5, col = "red")
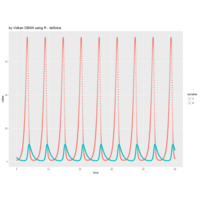
deSolve package
time <- seq(0, 50, by = 0.01)
# parameters: a named vector
parameters <- c(r = 2, k = 0.5, e = 0.1, d = 1)
# initial condition: a named vector
state <- c(V = 1, P = 3)
# R function to calculate the value of the derivatives at each time value
# Use the names of the variables as defined in the vectors above
lotkaVolterra <- function(t, state, parameters){
with(as.list(c(state, parameters)), {
dV = r * V - k * V * P
dP = e * k * V * P - d * P
return(list(c(dV, dP)))
})
}
## Integration with 'ode'
out <- ode(y = state, times = time, func = lotkaVolterra, parms = parameters)
## Ploting
out.df = as.data.frame(out) # required by ggplot: data object must be a data frame
library(reshape2)
out.m = melt(out.df, id.vars='time') # this makes plotting easier by puting all variables in a single column
p <- ggplot(out.m, aes(time, value, color = variable)) + geom_point()
p
igraph
g <‐ make_lattice(dimvector = c(5,5),
+ circular = FALSE)
plot(g,vertex.color=c("red", "darkslateblue","gold","lightblue","pink"))

igraph
> g<- make_tree(60, children=3)
> plot(g,vertex.color=c("red", "darkslateblue","gold","lightblue","pink"))

Sales Funnel with R . by means of https://www.r-bloggers.com/
library(dplyr)
library(ggplot2)
library(reshape2)
# creating a data samples
# content
df.content <- data.frame(content = c('main', 'ad landing',
'product 1', 'product 2', 'product 3', 'product 4',
'shopping cart',
'thank you page'),
step = c('awareness', 'awareness',
'interest', 'interest', 'interest', 'interest',
'desire',
'action'),
number = c(150000, 80000,
80000, 40000, 35000, 25000,
130000,
120000))
# customers
df.customers <- data.frame(content = c('new', 'engaged', 'loyal'),
step = c('new', 'engaged', 'loyal'),
number = c(25000, 40000, 55000))
# combining two data sets
df.all <- rbind(df.content, df.customers)
# calculating dummies, max and min values of X for plotting
df.all <- df.all %>%
group_by(step) %>%
mutate(totnum = sum(number)) %>%
ungroup() %>%
mutate(dum = (max(totnum) - totnum)/2,
maxx = totnum + dum,
minx = dum)
# data frame for plotting funnel lines
df.lines <- df.all %>%
select(step, maxx, minx) %>%
group_by(step) %>%
unique()
# data frame with dummies
df.dum <- df.all %>%
select(step, dum) %>%
unique() %>%
mutate(content = 'dummy',
number = dum) %>%
select(content, step, number)
# data frame with rates
conv <- df.all$totnum[df.all$step == 'action']
df.rates <- df.all %>%
select(step, totnum) %>%
group_by(step) %>%
unique() %>%
ungroup() %>%
mutate(prevnum = lag(totnum),
rate = ifelse(step == 'new' | step == 'engaged' | step == 'loyal',
round(totnum / conv, 3),
round(totnum / prevnum, 3))) %>%
select(step, rate)
df.rates <- na.omit(df.rates)
# creting final data frame
df.all <- df.all %>%
select(content, step, number)
df.all <- rbind(df.all, df.dum)
df.all <- df.all %>%
group_by(step) %>%
arrange(desc(content)) %>%
ungroup()
# calculating position of labels
df.all <- df.all %>%
group_by(step) %>%
mutate(pos = cumsum(number) - 0.5*number)
# defining order of steps
df.all$step <- factor(df.all$step, levels = c('loyal', 'engaged', 'new', 'action', 'desire', 'interest', 'awareness'))
list <- c(unique(as.character(df.all$content)))
df.all$content <- factor(df.all$content, levels = c('dummy', c(list)))
# creating custom palette with 'white' color for dummies
cols <- c("#ffffff", "#fec44f", "#fc9272", "#a1d99b", "#fee0d2", "#2ca25f",
"#8856a7", "#43a2ca", "#fdbb84", "#e34a33",
"#a6bddb", "#dd1c77", "#ffeda0", "#756bb1")
# plotting chart
ggplot() +
theme_minimal() +
coord_flip() +
scale_fill_manual(values=cols) +
geom_bar(data=df.all, aes(x=step, y=number, fill=content), stat="identity", width=1) +
geom_text(data=df.all[df.all$content!='dummy', ],
aes(x=step, y=pos, label=paste0(content, '-', number/1000, 'K')),
size=4, color='white', fontface="bold") +
geom_ribbon(data=df.lines, aes(x=step, ymax=max(maxx), ymin=maxx, group=1), fill='white') +
geom_line(data=df.lines, aes(x=step, y=maxx, group=1), color='darkred', size=4) +
geom_ribbon(data=df.lines, aes(x=step, ymax=minx, ymin=min(minx), group=1), fill='white') +
geom_line(data=df.lines, aes(x=step, y=minx, group=1), color='darkred', size=4) +
geom_text(data=df.rates, aes(x=step, y=(df.lines$minx[-1]), label=paste0(rate*100, '%')), hjust=1.2,
color='darkblue', fontface="bold") +
theme(legend.position='none', axis.ticks=element_blank(), axis.text.x=element_blank(),
axis.title.x=element_blank())

Sales Funnel with R . by means of https://www.r-bloggers.com/
library(tidyverse)
library(purrrlyr)
library(reshape2)
##### simulating the "real" data #####
set.seed(454)
df_raw <- data.frame(customer_id = paste0('id', sample(c(1:5000), replace = TRUE)),
date = as.POSIXct(rbeta(10000, 0.7, 10) * 10000000, origin = '2017-01-01', tz = "UTC"),
channel = paste0('channel_', sample(c(0:7), 10000, replace = TRUE, prob = c(0.2, 0.12, 0.03, 0.07, 0.15, 0.25, 0.1, 0.08))),
site_visit = 1) %>%
mutate(two_pages_visit = sample(c(0,1),
10000,
replace = TRUE,
prob = c(0.8, 0.2)),
product_page_visit = ifelse(two_pages_visit == 1,
sample(c(0, 1),
length(two_pages_visit[which(two_pages_visit == 1)]),
replace = TRUE, prob = c(0.75, 0.25)),
0),
add_to_cart = ifelse(product_page_visit == 1,
sample(c(0, 1),
length(product_page_visit[which(product_page_visit == 1)]),
replace = TRUE, prob = c(0.1, 0.9)),
0),
purchase = ifelse(add_to_cart == 1,
sample(c(0, 1),
length(add_to_cart[which(add_to_cart == 1)]),
replace = TRUE, prob = c(0.02, 0.98)),
0)) %>%
dmap_at(c('customer_id', 'channel'), as.character) %>%
arrange(date) %>%
mutate(session_id = row_number()) %>%
arrange(customer_id, session_id)
df_raw <- melt(df_raw, id.vars = c('customer_id', 'date', 'channel', 'session_id'), value.name = 'trigger', variable.name = 'event') %>%
filter(trigger == 1) %>%
select(-trigger) %>%
arrange(customer_id, date)
df_customers <- df_raw %>%
group_by(customer_id, event) %>%
filter(date == min(date)) %>%
ungroup()
sf_probs <- df_customers %>%
group_by(event) %>%
summarise(customers_on_step = n()) %>%
ungroup() %>%
mutate(sf_probs = round(customers_on_step / customers_on_step[event == 'site_visit'], 3),
sf_probs_step = round(customers_on_step / lag(customers_on_step), 3),
sf_probs_step = ifelse(is.na(sf_probs_step) == TRUE, 1, sf_probs_step),
sf_importance = 1 - sf_probs_step,
sf_importance_weighted = sf_importance / sum(sf_importance)
)
df_customers_plot <- df_customers %>%
group_by(event) %>%
arrange(channel) %>%
mutate(pl = row_number()) %>%
ungroup() %>%
mutate(pl_new = case_when(
event == 'two_pages_visit' ~ round((max(pl[event == 'site_visit']) - max(pl[event == 'two_pages_visit'])) / 2),
event == 'product_page_visit' ~ round((max(pl[event == 'site_visit']) - max(pl[event == 'product_page_visit'])) / 2),
event == 'add_to_cart' ~ round((max(pl[event == 'site_visit']) - max(pl[event == 'add_to_cart'])) / 2),
event == 'purchase' ~ round((max(pl[event == 'site_visit']) - max(pl[event == 'purchase'])) / 2),
TRUE ~ 0
),
pl = pl + pl_new)
df_customers_plot$event <- factor(df_customers_plot$event, levels = c('purchase',
'add_to_cart',
'product_page_visit',
'two_pages_visit',
'site_visit'
))
# color palette
cols <- c('#4e79a7', '#f28e2b', '#e15759', '#76b7b2', '#59a14f',
'#edc948', '#b07aa1', '#ff9da7', '#9c755f', '#bab0ac')
ggplot(df_customers_plot, aes(x = event, y = pl)) +
theme_minimal() +
scale_colour_manual(values = cols) +
coord_flip() +
geom_line(aes(group = customer_id, color = as.factor(channel)), size = 0.05) +
geom_text(data = sf_probs, aes(x = event, y = 1, label = paste0(sf_probs*100, '%')), size = 4, fontface = 'bold') +
guides(color = guide_legend(override.aes = list(size = 2))) +
theme(legend.position = 'bottom',
legend.direction = "horizontal",
panel.grid.major.x = element_blank(),
panel.grid.minor = element_blank(),
plot.title = element_text(size = 20, face = "bold", vjust = 2, color = 'black', lineheight = 0.8),
axis.title.y = element_text(size = 16, face = "bold"),
axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_text(size = 8, angle = 90, hjust = 0.5, vjust = 0.5, face = "plain")) + ggtitle("Sales Funnel visualization - all customers journeys")
ref:https://www.r-bloggers.com/marketing-multi-channel-attribution-model-based-on-sales-funnel-with-r/

naniar package
gg_miss_case(airquality)

PGRdup
GN1 <- GN1000[!grepl("^ICG", GN1000$DonorID), ]
GN1$DonorID <- NULL
GN2 <- GN1000[grepl("^ICG", GN1000$DonorID), ]
GN2 <- GN2[!grepl("S", GN2$DonorID), ]
GN2$NationalID <- NULL
GN1$SourceCountry <- toupper(GN1$SourceCountry)
GN2$SourceCountry <- toupper(GN2$SourceCountry)
GN1$SourceCountry <- gsub("UNITED STATES OF AMERICA", "USA", GN1$SourceCountry)
GN2$SourceCountry <- gsub("UNITED STATES OF AMERICA", "USA", GN2$SourceCountry)
# Specify as a vector the database fields to be used
GN1fields <- c("NationalID", "CollNo", "OtherID1", "OtherID2")
GN2fields <- c("DonorID", "CollNo", "OtherID1", "OtherID2")
# Clean the data
GN1[GN1fields] <- lapply(GN1[GN1fields], function(x) DataClean(x))
GN2[GN2fields] <- lapply(GN2[GN2fields], function(x) DataClean(x))
y1 <- list(c("Gujarat", "Dwarf"), c("Castle", "Cary"), c("Small", "Japan"),
c("Big", "Japan"), c("Mani", "Blanco"), c("Uganda", "Erect"),
c("Mota", "Company"))
y2 <- c("Dark", "Light", "Small", "Improved", "Punjab", "SAM")
y3 <- c("Local", "Bold", "Cary", "Mutant", "Runner", "Giant", "No.",
"Bunch", "Peanut")
GN1[GN1fields] <- lapply(GN1[GN1fields], function(x) MergeKW(x, y1, delim = c("space", "dash")))
GN1[GN1fields] <- lapply(GN1[GN1fields], function(x) MergePrefix(x, y2, delim = c("space", "dash")))
GN1[GN1fields] <- lapply(GN1[GN1fields], function(x) MergeSuffix(x, y3, delim = c("space", "dash")))
GN2[GN2fields] <- lapply(GN2[GN2fields], function(x) MergeKW(x, y1, delim = c("space", "dash")))
GN2[GN2fields] <- lapply(GN2[GN2fields], function(x) MergePrefix(x, y2, delim = c("space", "dash")))
GN2[GN2fields] <- lapply(GN2[GN2fields], function(x) MergeSuffix(x, y3, delim = c("space", "dash")))
# Remove duplicated DonorID records in GN2
GN2 <- GN2[!duplicated(GN2$DonorID), ]
# Generate KWIC index
GN1KWIC <- KWIC(GN1, GN1fields)
GN2KWIC <- KWIC(GN2, GN2fields)
# Specify the exceptions as a vector
exep <- c("A", "B", "BIG", "BOLD", "BUNCH", "C", "COMPANY", "CULTURE",
"DARK", "E", "EARLY", "EC", "ERECT", "EXOTIC", "FLESH", "GROUNDNUT",
"GUTHUKAI", "IMPROVED", "K", "KUTHUKADAL", "KUTHUKAI", "LARGE",
"LIGHT", "LOCAL", "OF", "OVERO", "P", "PEANUT", "PURPLE", "R",
"RED", "RUNNER", "S1", "SAM", "SMALL", "SPANISH", "TAN", "TYPE",
"U", "VALENCIA", "VIRGINIA", "WHITE")
# Specify the synsets as a list
syn <- list(c("CHANDRA", "AH114"), c("TG1", "VIKRAM"))
GNdupc <- ProbDup(kwic1 = GN1KWIC, kwic2 = GN2KWIC, method = "c",
excep = exep, fuzzy = TRUE, phonetic = TRUE,
encoding = "primary", semantic = TRUE, syn = syn)
GNdupcView <- ViewProbDup(GNdupc, GN1, GN2, "SourceCountry", "SourceCountry",
max.count = 30, select = c("INDIA", "USA"), order = "type",
main = "Groundnut Probable Duplicates")
library(gridExtra)
grid.arrange(GNdupcView$SummaryGrob)
ref:https://cran.r-project.org/web/packages/PGRdup/PGRdup.pdf
persp-- Perspective Plots
layout(matrix(1:9, ncol = 3, byrow = T))
> par(mar = c(0,0,0,0))
>
> for(i in seq(0, 360, length.out = 9)) {
+ persp(x = axis.vector,
+ y = axis.vector,
+ z = z.axis.vector.2,main=""+ theta = i, phi = 15,col = "springgreen", expand = 0.6, shade = 0.3) }
persp
> f.sugakuart.com <- function(a, b, x, y) {
+ a * exp(- (x - y)^2 / b)
+ }
>
> z.axis.vector.2 <- outer(X = axis.vector, Y = axis.vector, function(X,Y) f.sugakuart.com(a = 1, b = 1, x = X, y = Y))
>
> persp(x = axis.vector,
+ y = axis.vector,
+ z = z.axis.vector.2,main="",
+ theta = 100, phi = 30,col = "springgreen", expand = 0.6, shade = 0.3)

persp-
> f.sugakuart.com <- function(a, b, x, y) {
+ a * exp(- (x - y)^2 / b)
+ }
>
> z.axis.vector.2 <- outer(X = axis.vector, Y = axis.vector, function(X,Y) f.sugakuart.com(a = 1, b = 1, x = X, y = Y))
>
> persp(x = axis.vector,
+ y = axis.vector,
+ z = z.axis.vector.2,main="",
+ theta = 120, phi = 15,col = "springgreen", expand = 0.6, shade = 0.3)
>

grDevices
persp function
F<-function(x, y){
+ sqrt(cos(x)+sin(y))
> x <- y <- seq(-1, 1, length= 20)
> z <- outer(x, y, F)
> persp(x, y, zn",
+ zlab = "z",
+ theta = 30, phi = 15,
+ col = "springgreen", shade = 0.5)

lattice package
my.settings <- list(
+ par.main.text = list(font = 2, # make it bold
+ just = "left",
+ x = grid::unit(5, "mm")))
>
> xyplot(sin(1:200) ~ cos(1:200),
+ par.settings=my.settings,
+ main=" ", sub=" ",
+ type="l")
diagram
ref: http://www.rforscience.com/portfolio/solving-differential-equations-in-r-chapter-5/
Plot
require(shape)
par (mar = c(1, 1, 1, 1))
emptyplot()
mid <- c(0.5, 0.9)
r <- 0.8
dpi <- 0.18
GE <- getellipse (mid = mid, from = (3/2-dpi)*pi, to = (3/2 + dpi)*pi,
rx = r, ry = r)
plotcircle(mid = mid, from = (3/2-dpi)*pi, to = (3/2 + dpi)*pi,
lty = 1, lcol = "pink", r = r)
segments(mid[1], mid[2], mid[1], mid[2] - r, lty = 2)
nr <- nrow(GE) * 0.8
bob <- GE[nr, ]
segments(mid[1], mid[2], bob[1], bob[2], lty = 1, lwd = 2)
plotcircle(mid = mid, from = 3/2*pi, to = (3/2 + dpi*0.5)*pi,
lty = 1, lcol = "purple", r = r, arrow = TRUE,
arr.adj = 1, arr.type = "triangle", arr.length = 0.3)
filledellipse( mid = bob, col = greycol(100), rx1 = 0.035)
filledellipse( mid = mid - c(0, r), col = greycol(100, interval = c(0, 0.4)),
rx1 = 0.035)
filledellipse( mid = mid, col = "black", rx1 = 0.01)
xa <- 0.75
ya <- 0.7
dd <- 0.15
Arrows(xa, ya, xa, ya+dd, arr.type = "triangle", arr.length = 0.2)
Arrows(xa, ya, xa+dd, ya, arr.type = "triangle", arr.length = 0.2)
text(xa + dd/2, ya - dd/4, "x")
text(xa - dd/4, ya + dd/2, "y")
text(0.68, 0.45, " length L", adj = 0)
text(bob[1] + dd/3, bob[2], "m = 2", adj = 0)
ref:http://www.rforscience.com/portfolio/solving-differential-equations-in-r-chapter-4/
Plot
require(OceanView)
> require(shape)
> cols <- ramp.col(c( "lightblue1", "green"), n = 50)
> par(mar = c(0, 0, 0, 1))
> image2D(Hypsometry, col = cols, shade = 0.08, rasterImage = TRUE,
+ contour = list(levels = 0, draw = F), axes = FALSE, main="", xlab = ", ylab = "",
+ colkey = list(width = 0.3, length = 0.3, cex.axis = 0.5))
>

Plot3D package
> url <- "http://seamap.env.duke.edu/species/180524"
>
> require(plot3D)
> # terms of use: citation of OBIS-SEAMAP
>
> Mink <- read.csv("species_180524_points.csv") [, c
>
> # project on a grid
> nbins <- 200
> xm <- seq(-180, 180, length.out = nbins)
> ym <- seq(-90, 90, length.out = nbins)
> xy <- table(cut(Mink$longitude, xm),
+ cut(Mink$latitude, ym))
> xy [xy == 0] <- NA
> xmid <- 0.5*(xm[-1] + xm[-nbins])
> ymid <- 0.5*(ym[-1] + ym[-nbins])
>
> par(oma = c(2, 0, 0, 0))
> ImageOcean(col = ramp.col (c("lightblue", "darkblue")), shade = 0.1,
+ contour = list(levels = 0), NAcol = "grey", colkey = list (plot = FALSE),
+ main = " Minkwhale - OBIS seamap")
>
> image2D(x = xmid, y = ymid, z = xy, log = "c", add = TRUE,
+ col = jet2.col(100), NAcol = "transparent", clab = "count")
GA
y <- x <- seq(-10, 10, length=60)
> f <- function(x,y) { r <- sqrt(x^2+y^4); 10 * 2*sin(2*r)/r }
> z <- outer(x, y, f)
> persp3D(x, y, z, theta = 45,main="by Volkan OBAN using R - GA ", phi = 30, expand = 0.5
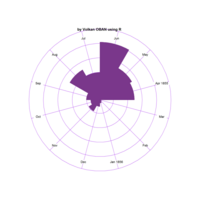
radialpie
> library(HistData)
Warning message:
package ‘HistData’ was built under R version 3.4.1
> library(plotrix)
> data = Nightingale[13:24,]
radial.pie

vipPlot
vioplot.singmann <- function(x, ..., range = 1.5, h = NULL, ylim = NULL, names = NULL,
horizontal = FALSE, col = NULL, border = "black", lty = 1, lwd = 1, rectCol = "black",
colMed = "white", pchMed = 19, at, add = FALSE, wex = 1, mark.outlier = TRUE,
pch.mean = 4, ids = NULL, drawRect = TRUE, yaxt = "s") {
# process multiple datas
datas <- list(x, ...)
n <- length(datas)
if (missing(at))
at <- 1:n
# pass 1 - calculate base range - estimate density setup parameters for
# density estimation
upper <- vector(mode = "numeric", length = n)
lower <- vector(mode = "numeric", length = n)
q1 <- vector(mode = "numeric", length = n)
q3 <- vector(mode = "numeric", length = n)
med <- vector(mode = "numeric", length = n)
base <- vector(mode = "list", length = n)
height <- vector(mode = "list", length = n)
outliers <- vector(mode = "list", length = n)
baserange <- c(Inf, -Inf)
# global args for sm.density function-call
args <- list(display = "none")
if (!(is.null(h)))
args <- c(args, h = h)
for (i in 1:n) {
data <- datas[[i]]
if (!is.null(ids))
names(data) <- ids
if (is.null(names(data)))
names(data) <- as.character(1:(length(data)))
# calculate plot parameters 1- and 3-quantile, median, IQR, upper- and
# lower-adjacent
data.min <- min(data)
data.max <- max(data)
q1[i] <- quantile(data, 0.25)
q3[i] <- quantile(data, 0.75)
med[i] <- median(data)
iqd <- q3[i] - q1[i]
upper[i] <- min(q3[i] + range * iqd, data.max)
lower[i] <- max(q1[i] - range * iqd, data.min)
# strategy: xmin = min(lower, data.min)) ymax = max(upper, data.max))
est.xlim <- c(min(lower[i], data.min), max(upper[i], data.max))
# estimate density curve
smout <- do.call("sm.density", c(list(data, xlim = est.xlim), args))
# calculate stretch factor the plots density heights is defined in range 0.0
# ... 0.5 we scale maximum estimated point to 0.4 per data
hscale <- 0.4/max(smout$estimate) * wex
# add density curve x,y pair to lists
base[[i]] <- smout$eval.points
height[[i]] <- smout$estimate * hscale
t <- range(base[[i]])
baserange[1] <- min(baserange[1], t[1])
baserange[2] <- max(baserange[2], t[2])
min.d <- boxplot.stats(data)[["stats"]][1]
max.d <- boxplot.stats(data)[["stats"]][5]
height[[i]] <- height[[i]][(base[[i]] > min.d) & (base[[i]] < max.d)]
height[[i]] <- c(height[[i]][1], height[[i]], height[[i]][length(height[[i]])])
base[[i]] <- base[[i]][(base[[i]] > min.d) & (base[[i]] < max.d)]
base[[i]] <- c(min.d, base[[i]], max.d)
outliers[[i]] <- list(data[(data < min.d) | (data > max.d)], names(data[(data <
min.d) | (data > max.d)]))
# calculate min,max base ranges
}
# pass 2 - plot graphics setup parameters for plot
if (!add) {
xlim <- if (n == 1)
at + c(-0.5, 0.5) else range(at) + min(diff(at))/2 * c(-1, 1)
if (is.null(ylim)) {
ylim <- baserange
}
}
if (is.null(names)) {
label <- 1:n
} else {
label <- names
}
boxwidth <- 0.05 * wex
# setup plot
if (!add)
plot.new()
if (!horizontal) {
if (!add) {
plot.window(xlim = xlim, ylim = ylim)
axis(2)
axis(1, at = at, label = label)
}
box()
for (i in 1:n) {
# plot left/right density curve
polygon(c(at[i] - height[[i]], rev(at[i] + height[[i]])), c(base[[i]],
rev(base[[i]])), col = col, border = border, lty = lty, lwd = lwd)
if (drawRect) {
# browser() plot IQR
boxplot(datas[[i]], at = at[i], add = TRUE, yaxt = yaxt, pars = list(boxwex = 0.6 *
wex, outpch = if (mark.outlier) "" else 1))
if ((length(outliers[[i]][[1]]) > 0) & mark.outlier)
text(rep(at[i], length(outliers[[i]][[1]])), outliers[[i]][[1]],
labels = outliers[[i]][[2]])
# lines( at[c( i, i)], c(lower[i], upper[i]) ,lwd=lwd, lty=lty) plot 50% KI
# box rect( at[i]-boxwidth/2, q1[i], at[i]+boxwidth/2, q3[i], col=rectCol)
# plot median point points( at[i], med[i], pch=pchMed, col=colMed )
}
points(at[i], mean(datas[[i]]), pch = pch.mean, cex = 1.3)
}
} else {
if (!add) {
plot.window(xlim = ylim, ylim = xlim)
axis(1)
axis(2, at = at, label = label)
}
box()
for (i in 1:n) {
# plot left/right density curve
polygon(c(base[[i]], rev(base[[i]])), c(at[i] - height[[i]], rev(at[i] +
height[[i]])), col = col, border = border, lty = lty, lwd = lwd)
if (drawRect) {
# plot IQR
boxplot(datas[[i]], yaxt = yaxt, at = at[i], add = TRUE, pars = list(boxwex = 0.8 *
wex, outpch = if (mark.outlier) "" else 1))
if ((length(outliers[[i]][[1]]) > 0) & mark.outlier)
text(rep(at[i], length(outliers[[i]][[1]])), outliers[[i]][[1]],
labels = outliers[[i]][[2]])
# lines( at[c( i, i)], c(lower[i], upper[i]) ,lwd=lwd, lty=lty)
}
points(at[i], mean(datas[[i]]), pch = pch.mean, cex = 1.3)
}
}
invisible(list(upper = upper, lower = lower, median = med, q1 = q1, q3 = q3))
}
# plot
par(cex.main = 1.5, mar = c(5, 6, 4, 5) + 0.1, mgp = c(3.5, 1, 0), cex.lab = 1.5,
font.lab = 2, cex.axis = 1.3, bty = "n", las = 1)
x <- c(1, 2, 3, 4)
plot(x, c(-10, -10, -10, -10), type = "p", ylab = " ", xlab = " ", cex = 1.5,
ylim = c(0.3, 0.6), xlim = c(1, 4), lwd = 2, pch = 5, axes = F, main = " ")
axis(1, at = c(1.5, 2.5, 3.5), labels = c("HF", "LF", "VLF"))
axis(2, pos = 1.1)
mtext("Word Frequency", side = 1, line = 3, cex = 1.5, font = 2)
par(las = 0)
mtext("Group Mean M", side = 2, line = 2.9, cex = 1.5, font = 2)
x <- c(1.5, 2.5, 3.5)
vioplot.singmann(RT.hf.sp, RT.lf.sp, RT.vlf.sp, add = TRUE, mark.outlier = FALSE,
at = c(1.5, 2.5, 3.5), wex = 0.4, yaxt = "n")
vioplot.singmann(RT.hf.ac, RT.lf.ac, RT.vlf.ac, add = TRUE, mark.outlier = FALSE,
at = c(1.5, 2.5, 3.5), wex = 0.4, col = "grey", border = "grey", rectCol = "grey",
colMed = "grey", yaxt = "n")
text(2.5, 0.35, "Speed", cex = 1.4, font = 1, adj = 0.5)
text(2.5, 0.58, "Accuracy", cex = 1.4, font = 1, col = "grey", adj = 0.5)
ref:http://shinyapps.org/apps/RGraphCompendium/index.php

arulesViz
> data(Groceries)
> rules <- apriori(Groceries, parameter=list(support=0.005, confidence=0.5))
> plot(rules, method="grouped")
ref:http://www.ekonlab.com/?p=835

ggformula
gf_point(price~carat| color ~ clarity, data=diamonds, alpha=0.2) %>% gf_lm()

ggformula
ggplot(data = iris, aes(sample = Sepal.Length)) +
+ geom_qq() +
+ stat_qqline( alpha = 0.7, color = "red", linetype = "dashed") +
+ facet_wrap(~Species)

ggformula
> D <- expand.grid(x = 1:10, y=1:10)
> D$angle <- runif(100, 0, 2*pi)
> D$speed <- runif(100, 0, sqrt(0.1 * D$x))
> gf_point(y ~ x, data = D) %>%
+ gf_spoke(y ~ x, angle = ~angle, radius = 0.5)
> gf_point(y ~ x, data = D) %>%
+ gf_spoke(y ~ x, angle = ~angle, radius = ~speed)
ggformula
if (require(weatherData) & require(dplyr)) {
+ Temps <- NewYork2013 %>% mutate(city = "NYC") %>%
+ bind_rows(Mumbai2013 %>% mutate(city = "Mumbai")) %>%
+ bind_rows(London2013 %>% mutate(city = "London")) %>%
+ mutate(date = lubridate::date(Time),
+ month = lubridate::month(Time)) %>%
+ group_by(city, date) %>%
+ summarise(
+ hi = max(Temperature, na.rm = TRUE),
+ lo = min(Temperature, na.rm = TRUE),
+ mid = (hi + lo)/2
+ )
+ gf_ribbon(lo + hi ~ date, data = Temps, fill = ~city, alpha = 0.4) %>%
+ gf_theme(theme = theme_minimal())
+ gf_linerange(lo + hi ~ date | city ~ ., color = ~mid, data = Temps) %>%
+ gf_refine(scale_colour_gradientn(colors = rev(rainbow(5))))
+ gf_ribbon(lo + hi ~ date | city ~ ., data = Temps)
+ # Chaining in the data
+ Temps %>% gf_ribbon(lo + hi ~ date, alpha = 0.4) %>% gf_facet_grid(city ~ .)
+ }

ggformula
gf_dotplot(~ Sepal.Length, fill = ~Species, data = iris)

geofacet
> ggplot(eu_gdp, aes(year, gdp_pc)) +
+ geom_line(color = "steelblue") +
+ geom_hline(yintercept = 100, linetype = 2) +
+ facet_geo(~ name, grid = "eu_grid1") +
+ scale_x_continuous(labels = function(x) paste0("'", substr(x, 3, 4))) +
+ ylab("GDP Per Capita") +
+ theme_bw()

geofacet
> library(geofacet)
Warning message:
package ‘geofacet’ was built under R version 3.4.1
> library(ggplot2)
> # barchart of state rankings in various categories
> ggplot(state_ranks, aes(variable, rank, fill = variable)) +
+ geom_col() +
+ coord_flip() +
+ facet_geo(~ state) +
+ theme_bw()
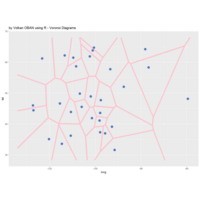
Voronoi Diagrams
> set.seed(105)
> long<-rnorm(30,-100,18)
> lat<-rnorm(30,49,12)
> df <- data.frame(lat,long)
>
> library(deldir)
> library(ggplot2)
>
> #This creates the voronoi line segments
> voronoi <- deldir(df$long, df$lat)
>
> #Now we can make a plot
> ggplot(data=df, aes(x=long,y=lat)) +
+ #Plot the voronoi lines
+ geom_segment(
+ aes(x = x1, y = y1, xend = x2, yend = y2),
+ size = 2,
+ data = voronoi$dirsgs,
+ linetype = 1,
+ color= "pink") +
+ #Plot the points
+ geom_point(
+ fill=rgb(70,130,180,255,maxColorValue=255),
+ pch=21,
+ size = 4,
+ color="purple")

cartogram
> library(maptools)
> library(cartogram)
> library(rgdal)
> data(wrld_simpl)
> afr <- spTransform(wrld_simpl[wrld_simpl$REGION==2 & wrld_simpl$POP2005 > 0,],
+ CRS("+init=epsg:3395"))
> par(mfcol=c(1,2))
> plot(afr)
> plot(cartogram(afr, "POP2005", 3))

tripack-k-means and voronoi diagrams
set.seed(1)
pts <- cbind(X=rnorm(500,rep(seq(1,9,by=2)/10,100),.022),Y=rnorm(500,.5,.15))
km1 <- kmeans(pts, centers=5, nstart = 1, algorithm = "Lloyd")
There were 19 warnings (use warnings() to see them)
> library(tripack)
> library(RColorBrewer)
> CL5 <- brewer.pal(5, "Pastel1")
> V <- voronoi.mosaic(km1$centers[,1],km1$centers[,2])
> P <- voronoi.polygons(V)
> plot(pts,pch=19,xlim=0:1,ylim=0:1,xlab="",ylab="",col=CL5[km1$cluster])
> points(km1$centers[,1],km1$centers[,2],pch=3,cex=1.5,lwd=2)
> plot(V,add=TRUE)
ref:http://freakonometrics.hypotheses.org

scatterplot3d
data(Mishkin )
ref: ref: Visualizing Complex Data Using R by N.D. Lewis

tm and wordcloud
data(SOTU)# contains the text of the Presidential addresses.
> # we only want the words so we remove punctuation
> text <- tm_map(SOTU, removePunctuation)
> text <- tm_map(text, function(x)removeWords (x,stopwords()))
> # put cleaned data in appropriate format
> tdm <- TermDocumentMatrix(text)
> m <- as.matrix(tdm)
> v <- sort(rowSums(m),decreasing=TRUE)
> d <- data.frame(word = names(v),freq=v)
> par(bg="purple4")# set background color
> wordcloud(d$word,d$freq, random.order=FALSE,min.freq=6 ,
color="navajowhite")
ref:Visualizing Complex Data Using R
by N.D. Lewis
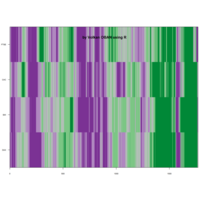
mvtsplot
ref: Visualizing Complex Data Using R
by N.D. Lewis
mvtsplot
> library(datasets)
> library(mvtsplot)
>D <- diff(EuStockMarkets ,90)
>mvtsplot(D,,norm ="internal", levels = 4,margin=FALSE)
spineplot
> r1 = c (7.9, 67.6, 28.3, 53.6)
> r2 = c (4.4, 54.5, 29.9, 57.6)
> r3 = c (10.2, 50, 27.7, 53.4)
> r4 = c (2.5, 35.3, 22.2, 47)
> r5 = c (8.5, 46.3, 32.2, 50)
> data <- as.table(rbind(r1,r2,r3,r4,r5))
> dimnames(data) <- list(x = c("volkan","oban", "V","O","VO"), R_spineplot = c("A (< 10)","B (<10)", "A (> 10)","B(> 10)"))
> spineplot(data)

Plot
> set.seed(345)
> Sector <- rep(c("S01","S02","S03","S04","S05","S06","S07"),times=7)
> Year <- as.numeric(rep(c("1950","1960","1970","1980","1990","2000","2010"),each=7))
> Value <- runif(49, 10, 100)
> data <- data.frame(Sector,Year,Value)
> ggplot(data, aes(x=Year, y=Value, fill=Sector)) +
+ geom_area(colour="black", size=.25, alpha=.4) + scale_fill_brewer(palette="Spectral", breaks=rev(levels(data$Sector))

plot3D
rect3D(x0 = 0.02, y0 = 0.25, z0 = 0.03, x1 = 1, z1 = 5,
+ ylim = c(0, 1), bty = "g", facets = TRUE",
+ border = "purple", col ="#7570B3", alpha=0.5,
+ lwd = 2, phi = 20)

Plot3D package
> data(iris)
> x <- sep.l <- iris$Sepal.Length
> y <- pet.l <- iris$Petal.Length
> z <- sep.w <- iris$Sepal.Width
> library(plot3D)
scatter3D(x, y, z, phi = 0, bty = "g", pch = 20, cex = 0.5)
> text3D(x, y, z, labels = rownames(iris), add = TRUE, colkey = FALSE, cex = 0.5)
ref: http://www.sthda.com

Plot3D package
> data(iris)
> x <- sep.l <- iris$Sepal.Length
> y <- pet.l <- iris$Petal.Length
> z <- sep.w <- iris$Sepal.Width
> library(plot3D)
Warning message:
package ‘plot3D’ was built under R version 3.4.1
> scatter3D(x, y, z, phi = 0, bty = "g", type = "b",
+ ticktype = "detailed", pch = 20,
+ cex = c(0.5, 1, 1.5))
ref:http://www.sthda.com

ggplot2
> y <- matrix(rnorm(500), 100, 5, dimnames=list(paste("g", 1:100, sep=""), paste("VO", 1:5, sep="")))
> y <- data.frame(Position=1:length(y[,1]), y)
>
> df <- melt(y, id.vars=c("Position"), variable.name = "VO", value.name="Values")
> p <- ggplot(df, aes(Position, Values)) + geom_line(aes(color=VO)) + facet_wrap(~VO, ncol=1)
> print(p)
> ggplot(df, aes(VO, Values, fill=VO)) + geom_boxplot()
>

ggplot2
> p <- ggplot(iris, aes(Sepal.Length, Sepal.Width)) +
+ geom_line(aes(color=Species), size=1) +
+ facet_wrap(~Species, ncol=1)
> p
> p
DATA ART with R
> theta = seq(0, 2*pi, length = 300)
> x = cos(theta)
> y = sin(theta)
>
> # set graphical parameters
> op = par(bg = "black", mar = rep(0.5, 4))
>
> # plot
> plot(x, y, type = 'n')
> segments(rep(0, 299), rep(0, 299), x[1:299] * runif(299, 0.5),
+ y[1:299] * runif(299, 0.7),
+ col = hsv(runif(95, 0.75, 0.85), 1, 1, runif(299, 0.5)),
+ lwd = 4*runif(299))
>
> # signature
> legend("topright", legend = "", bty = "n", text.col = "white")
Plot
> dat <- read.table(text = "A B C D E F G
+ 1 480 780 431 295 670 360 190
+ 2 720 350 377 255 340 615 345
+ 3 460 480 179 560 60 735 1260
+ 4 220 240 876 789 820 100 75", header = TRUE)
>
> library(reshape2)
>
> dat$row <- seq_len(nrow(dat))
> dat2 <- melt(dat, id.vars = "row")
>
> library(ggplot2)
Attaching package: ‘ggplot2’
The following objects are masked _by_ ‘.GlobalEnv’:
is.facet, midwest
>
> ggplot(dat2, aes(x=variable, y=value, fill=row)) +
+ geom_bar(stat="identity") +
+ xlab("\nType") +
+ ylab("Time\n") +
+ guides(fill=FALSE) +
+ theme_bw()

stripchart
set.seed(1); A <- sample(0:10, 100, replace = TRUE)
stripchart(A, method = "stack", offset = .5, at = .15, pch = 19,
main = "Dotplot of Random Values", xlab = "Random Values")

Plot
-
ref:Graphing Data with R.

FFtree
# Create FFTrees of the heart disease data
heart.fft <- FFTrees(formula = diagnosis ~.,
data = heartdisease)
# Visualise the tree
plot(heart.fft,
main = "Heart Disease Diagnosis",
decision.labels = c("Absent", "Present"))

FFtree
> heart.fft <- FFTrees(formula = diagnosis ~., data = heartdisease)
heart.fft
# Plot the best tree
plot(heart.fft)

ggalt-hrbrthemes
> library(hrbrthemes)
> library(ggalt)
> library(tidyverse)
> sports <- read_tsv("https://github.com/halhen/viz-pub/raw/master/sports-time-of-day/activity.tsv")
Parsed with column specification:
cols(
activity = col_character(),
time = col_double(),
p = col_double()
)
>
> sports %>%
+ group_by(activity) %>%
+ filter(max(p) > 3e-04,
+ !grepl('n\\.e\\.c', activity)) %>%
+ arrange(time) %>%
+ mutate(p_peak = p / max(p),
+ p_smooth = (lag(p_peak) + p_peak + lead(p_peak)) / 3,
+ p_smooth = coalesce(p_smooth, p_peak)) %>%
+ ungroup() %>%
+ do({
+ rbind(.,
+ filter(., time == 0) %>%
+ mutate(time = 24*60))
+ }) %>%
+ mutate(time = ifelse(time < 3 * 60, time + 24 * 60, time)) %>%
+ mutate(activity = reorder(activity, p_peak, FUN=which.max)) %>%
+ arrange(activity) %>%
+ mutate(activity.f = reorder(as.character(activity), desc(activity))) -> sports
>
> sports <- mutate(sports, time2 = time/60)
>
> ggplot(sports, aes(time2, p_smooth)) +
+ geom_horizon(bandwidth=0.1) +
+ facet_grid(activity.f~.) +
+ scale_x_continuous(expand=c(0,0), breaks=seq(from = 3, to = 27, by = 3), labels = function(x) {sprintf("%02d:00", as.integer(x %% 24))}) +
+ viridis::scale_fill_viridis(name = "Activity relative to peak", discrete=TRUE,
+ labels=scales::percent(seq(0, 1, 0.1)+0.1)) +
+ labs(x=NULL, y=NULL, title="by Volkan OBAN using R - ggalt and hrbrthemes \n \n Peak time of day for sports and leisure",
+ subtitle="Number of participants throughout the day compared to peak popularity.") +
+ theme_ipsum_rc(grid="") +
+ theme(panel.spacing.y=unit(-0.05, "lines")) +
+ theme(strip.text.y = element_text(hjust=0, angle=360)) +
+ theme(axis.text.y=element_blank())

dumbbell plot
library(ggplot2) # devtools::install_github("hadley/ggplot2")
library(ggalt) # devtools::install_github("hrbrmstr/ggalt")
library(dplyr) # for data_frame() & arrange()
# I'm not crazy enough to input all the data; this will have to do for the example
df <- data_frame(country=c("Germany", "France", "Vietnam", "Japan", "Poland", "Lebanon",
"Australia", "SouthnKorea", "Canada", "Spain", "Italy", "Peru",
"U.S.", "UK", "Mexico", "Chile", "China", "India"),
ages_35=c(0.39, 0.42, 0.49, 0.43, 0.51, 0.57,
0.60, 0.45, 0.65, 0.57, 0.57, 0.65,
0.63, 0.59, 0.67, 0.75, 0.52, 0.48),
ages_18_to_34=c(0.81, 0.83, 0.86, 0.78, 0.86, 0.90,
0.91, 0.75, 0.93, 0.85, 0.83, 0.91,
0.89, 0.84, 0.90, 0.96, 0.73, 0.69),
diff=sprintf("+%d", as.integer((ages_18_to_34-ages_35)*100)))
# we want to keep the order in the plot, so we use a factor for country
df <- arrange(df, desc(diff))
df$country <- factor(df$country, levels=rev(df$country))
# we only want the first line values with "%" symbols (to avoid chart junk)
# quick hack; there is a more efficient way to do this
percent_first <- function(x) {
x <- sprintf("%d%%", round(x*100))
x[2:length(x)] <- sub("%$", "", x[2:length(x)])
x
}
gg <- ggplot()
# doing this vs y axis major grid line
gg <- gg + geom_segment(data=df, aes(y=country, yend=country, x=0, xend=1), color="#b2b2b2", size=0.15)
# dum…dum…dum!bell
gg <- gg + geom_dumbbell(data=df, aes(y=country, x=ages_35, xend=ages_18_to_34),
size=1.5, color="#b2b2b2", point.size.l=3, point.size.r=3,
point.colour.l="#9fb059", point.colour.r="#edae52")
# text below points
gg <- gg + geom_text(data=filter(df, country=="Germany"),
aes(x=ages_35, y=country, label="Ages 35+"),
color="#9fb059", size=3, vjust=-2, fontface="bold", family="Calibri")
gg <- gg + geom_text(data=filter(df, country=="Germany"),
aes(x=ages_18_to_34, y=country, label="Ages 18-34"),
color="#edae52", size=3, vjust=-2, fontface="bold", family="Calibri")
# text above points
gg <- gg + geom_text(data=df, aes(x=ages_35, y=country, label=percent_first(ages_35)),
color="#9fb059", size=2.75, vjust=2.5, family="Calibri")
gg <- gg + geom_text(data=df, color="#edae52", size=2.75, vjust=2.5, family="Calibri",
aes(x=ages_18_to_34, y=country, label=percent_first(ages_18_to_34)))
# difference column
gg <- gg + geom_rect(data=df, aes(xmin=1.05, xmax=1.175, ymin=-Inf, ymax=Inf), fill="#efefe3")
gg <- gg + geom_text(data=df, aes(label=diff, y=country, x=1.1125), fontface="bold", size=3, family="Calibri")
gg <- gg + geom_text(data=filter(df, country=="Germany"), aes(x=1.1125, y=country, label="DIFF"),
color="#7a7d7e", size=3.1, vjust=-2, fontface="bold", family="Calibri")
gg <- gg + scale_x_continuous(expand=c(0,0), limits=c(0, 1.175))
gg <- gg + scale_y_discrete(expand=c(0.075,0))
gg <- gg + labs(x=NULL, y=NULL, title="The social media age gap",
subtitle="Adult internet users or reported smartphone owners whonuse social networking sites",
caption="Source: Pew Research Center, Spring 2015 Global Attitudes Survey. Q74")
gg <- gg + theme_bw(base_family="Calibri")
gg <- gg + theme(panel.grid.major=element_blank())
gg <- gg + theme(panel.grid.minor=element_blank())
gg <- gg + theme(panel.border=element_blank())
gg <- gg + theme(axis.ticks=element_blank())
gg <- gg + theme(axis.text.x=element_blank())
gg <- gg + theme(plot.title=element_text(face="bold"))
gg <- gg + theme(plot.subtitle=element_text(face="italic", size=9, margin=margin(b=12)))
gg <- gg + theme(plot.caption=element_text(size=7, margin=margin(t=12), color="#7a7d7e"))
gg

ggjoy
ref :http://lenkiefer.com/2017/08/03/joyswarm
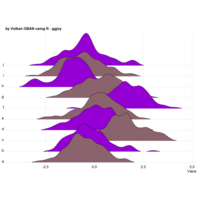
ggjoy
set.seed(123)
dt<- data.frame('label'=rep(letters[1:10], each=100),
'value'=as.vector(mapply(rnorm, rep(100, 10), rnorm(10), SIMPLIFY=TRUE)),
'rank'=rep(1:5, each=100, times=20))
ggplot(dt, aes(x=value, y=label, fill=label)) +
+ geom_joy(scale=3, rel_min_height=0.01) +
+ scale_fill_manual(values=rep(c('pink4', 'darkviolet'), length(unique(joy$label))/2)) +
+ scale_y_discrete(expand = c(0.01, 0)) +
+ xlab('Value') +
+ theme_joy() +
+ theme(axis.title.y = element_blank(),
+ legend.position='none')

gjoy
> p1 = ggtree(tr) %<+% d1 +
+ geom_tippoint(aes(color=location), size=5) +
+ geom_tiplab(offset=-0.01, hjust=0.5, colour="white", size=3, fontface="bold") + ggtitle("by Volkan OBAN using R - ggjoy") +
+ scale_colour_manual(values = c("purple", "pink", "yellow")) +
+ scale_fill_manual(values = c("purple", "pink", "yellow"))
>
> facet_plot(p1, panel="Joy Plot", data=d4, geom_joy,
+ mapping = aes(x=val, group=label, fill=location), colour="grey40", lwd=0.3)
ref:https://stackoverflow.com/questions/45384281/ggjoy-facet-with-ggtree

gjoy
> require(ggtree)
> require(ggstance)
> # generate tree
> tr <- rtree(30)
>
> # create simple ggtree object with tip labels
> p <- ggtree(tr) + geom_tiplab(offset = 0.02)
>
> # Generate categorical data for each "species"
> d1 <- data.frame(id=tr$tip.label, location=sample(c("GZ", "HK", "CZ"), 30, replace=TRUE))
>
> #Plot the categorical data as colored points on the tree tips
> p1 <- p %<+% d1 + geom_tippoint(aes(color=location))
>
> # Generate distribution of points for each species
> d4 = data.frame(id=rep(tr$tip.label, each=20),
+ val=as.vector(sapply(1:30, function(i)
+ rnorm(20, mean=i)))
+ )
>
> require(ggjoy)
>
> ggplot(d4, aes(x = val, y = id)) +
+ geom_joy(scale = 2, rel_min_height=0.03) +
+ scale_y_discrete(expand = c(0.01, 0)) + theme_joy() + ggtitle("by Volkan OBAN using R - ggjoy")
Picking joint bandwidth of 0.439
> p <- ggtree(tr) + geom_tiplab(offset = 0.02);p1 <- p %<+% d1 + geom_tippoint(aes(color=location));facet_plot(p1, panel="Joy Plot", data=d4, geom_joy,
+ mapping = aes(x=val, group=label, fill=location), colour="grey50", lwd=0.3)

ggjoy
> set.seed(1234)
> pois_data <- data.frame(mean = rep(1:5, each = 10))
> pois_data$group <- factor(pois_data$mean, levels=5:1)
> pois_data$value <- rpois(nrow(pois_data), pois_data$mean)
>
> # make plot
> ggplot(pois_data, aes(x = value, y = group, group = group)) +
+ geom_joy2(aes(fill = group), stat = "binline", binwidth = 1, scale = 0.95) +
+ geom_text(stat = "bin",
+ aes(y = group + 0.95*(..count../max(..count..)),
+ label = ifelse(..count..>0, ..count.., "")),
+ vjust = 1.4, size = 3, color = "white", binwidth = 1) +
+ scale_x_continuous(breaks = c(0:12), limits = c(-.5, 13), expand = c(0, 0),
+ name = "random value") +
+ scale_y_discrete(expand = c(0.01, 0), name = "Poisson mean",
+ labels = c("5.0", "4.0", "3.0", "2.0", "1.0")) +
+ scale_fill_cyclical(values = c("#0000B0", "#7070D0")) +
+ labs(title = " Poisson random samples with different means",
+ subtitle = "sample size n=10") +
+ guides(y = "none") +
+ theme_joy(grid = FALSE) +
+ theme(axis.title.x = element_text(hjust = 0.5),
+ axis.title.y = element_text(hjust = 0.5))
ref: https://cran.r-project.org/web/packages/ggjoy/vignettes/gallery.html
ggjoy
> library(ggplot2movies)
> ggplot(movies[movies$year>1912,], aes(x = length, y = year, group = year)) +
+ geom_joy(scale = 10, size = 0.25, rel_min_height = 0.03) +
+ theme_joy() +
+ scale_x_continuous(limits=c(1, 200), expand = c(0.01, 0)) +
+ scale_y_reverse(breaks=c(2000, 1980, 1960, 1940, 1920, 1900), expand = c(0.01, 0))
ggjoy
ggplot(diamonds, aes(x = price, y = cut, fill = cut)) +
+ geom_joy(scale = 4) +
+ scale_fill_cyclical(values = c("purple", "pink"))

ggjoy
> library(ggjoy)
Warning message:
package ‘ggjoy’ was built under R version 3.4.1
>
> ggplot(diamonds, aes(x = price, y = cut)) +
+ geom_joy(scale = 4) + theme_joy() +
+ scale_y_discrete(expand = c(0.01, 0)) + # will generally have to set the `expand` option
+ scale_x_continuous(expand = c(0, 0))

cowplot
a<- qplot(color, price/carat, data = diamonds, geom = "jitter", alpha = I(1/15))
ggdraw(a) +
+ draw_plot_label("R - Data Visualization-data(diamonds)", size = 12) +
+ draw_label("", angle = 25, size = 50, alpha = .7)
cowplot
a<-ggplot(data=diamonds,aes(x=price, group=cut, fill=cut)) +
geom_density(adjust=1.5, position="fill")
ggdraw(a) +
+ draw_plot_label("Data Science & Analytics", size = 8) +
+ draw_label("", angle = 45, size = 40, alpha = .6)
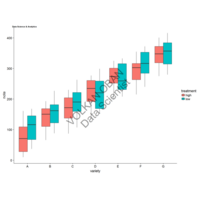
cowplot
ggdraw

lattice package
df <- data.frame(expand.grid(1:100,1:100),rep(10,1000)) ;colnames(df) <- c("x","y","z"); wireframe(z~x*y,df,colorkey=TRUE,drape=TRUE);wireframe(z~x*y,df,main="",color="",drape=TRUE, zlim=c(0,24))
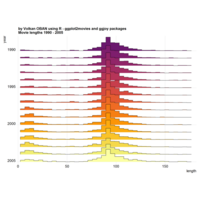
ggjoy
> require(ggplot2movies)
> require(viridis)
> ggplot(movies[movies$year>1989,], aes(x = length, y = year, fill = factor(year))) +
+ stat_binline(scale = 1.9, bins = 40) +
+ theme_joy() + theme(legend.position = "none") +
+ scale_x_continuous(limits = c(1, 180), expand = c(0.01, 0)) +
+ scale_y_reverse(expand = c(0.01, 0)) +
+ scale_fill_viridis(begin = 0.3, discrete = TRUE, option = "B") +
+ labs(title = " Movie lengths 1990 - 2005")

ggjoy
ggplot(iris, aes(x = Sepal.Length, y = Species, group = Species)) +
+ geom_joy(rel_min_height = 0.005) +
+ scale_y_discrete(expand = c(0.01, 0)) +
+ scale_x_continuous(expand = c(0.01, 0)) +
+ theme_joy()
persp-- Perspective Plots
cone <- function(x, y){ sqrt(x*cos(x^2)+sin(y)) } ;x <- y <- seq(-1, 1, length= 50); z <- outer(x, y, cone); persp(x, y, z, main="" ,col="pink")
GA
y <- x <- seq(-10, 10, length=60)
f <- function(x,y) { r <- sqrt(x^2+y^2); 10 * sin(r)/r }
z <- outer(x, y, f)
persp3D(x, y, z, color.palette = heat.colors, phi = 30, theta = 225,
box = TRUE, border = NA, shade = .4)

lattice package
df <- data.frame(expand.grid(1:10,1:10),rep(10,100)) ;colnames(df) <- c("x","y","z"); wireframe(z~x*y,df,colorkey=TRUE,drape=TRUE);wireframe(z~x*y,df,main="",colorkey=TRUE,drape=TRUE, zlim=c(0,24))
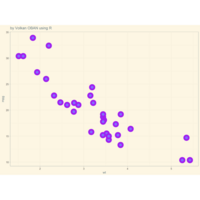
ggplot2
gplot(mtcars, aes(wt, mpg)) +
+ geom_point(shape = 21, colour = "purple", fill = "slateblue1", size = 5, stroke = 5) + theme_solarized() +
+ scale_colour_solarized("blue")

ggplot2
library("tidyverse")
library("forcats")
library(ggthemes)
rincome_plot <-
gss_cat %>%
ggplot(aes(rincome)) +
geom_bar()
rincome_plot
gss_cat %>%
filter(!denom %in% c("No answer", "Other", "Don't know", "Not applicable",
"No denomination")) %>%
count(relig)
gss_cat %>%
count(relig, denom) %>%
ggplot(aes(x = relig, y = denom, size = n)) +
geom_point() +
theme(axis.text.x = element_text(angle = 90) + theme_igray()
ref:https://jrnold.github.io/e4qf/factors.html
ggplot2
> dataframe <- tibble(
+ x = rnorm(10000),
+ y = rnorm(10000) )
ggplot(dataframe , aes(x, y)) +
+ geom_hex() +
+ scale_fill_gradient(low = "thistle2", high = "purple") +
+ coord_fixed()

ggplot2
> ggplot(mpg, aes(displ, hwy, colour = class)) +
+ geom_point(aes(colour = class)) +
+ geom_smooth(method = "lm", se = FALSE) +
+ labs(
+ title = "Fuel efficiency generally decreases with engine size",
+ subtitle = "Subcompact cars show the greatest sensitivity to engine size",
+ caption = "Data from fueleconomy.gov"
+ )
ggplot2
> library(gapminder)
Warning message:
package ‘gapminder’ was built under R version 3.4.1
> lifeExp ~ poly(year, 2)
lifeExp ~ poly(year, 2)
> country_model <- function(df) {
+ lm(lifeExp ~ poly(year - median(year), 2), data = df)
+ }
>
> by_country <- gapminder %>%
+ group_by(country, continent) %>%
+ nest()
>
> by_country <- by_country %>%
+ mutate(model = map(data, country_model))
> by_country <- by_country %>%
+ mutate(
+ resids = map2(data, model, add_residuals)
+ )
> by_country
unnest(by_country, resids) %>%
+ ggplot(aes(year, resid)) +
+ geom_line(aes(group = country), alpha = 1 / 3) +
+ geom_smooth(se = FALSE)

treemap
World population 2014
lattice package
> params.grid.length <- 20
> params.alpha.list <- seq(0.3, 0.6, length = params.grid.length)
> params.beta.list <- seq(1,9, length = params.grid.length)
> z <- matrix(ncol = params.grid.length, nrow = params.grid.length)
>
> # Loop through and calculate negative log likelihood at defined values in grid
> for (i in 1:length(params.alpha.list )){
+ for (ii in 1:length(params.beta.list)){
+ alpha <- params.alpha.list[i]
+ beta <- params.beta.list[ii]
+ y <- 0.5 + (1 - 0.5 - 0.025)* pweibull(resp.frame$x, beta, alpha)
+ negLog <- -sum(resp.frame$ny * log(y) + (resp.frame$num.tr - resp.frame$ny) * log(1 - y) ) # Negative log likelihood
+ z[i,ii] <- negLog
+ }
+ }
>
> # Need to generate stacked list of values to pass to wireframe in lattice
> params.alpha.wireList <- rep(unique(params.alpha.list),params.grid.length)
> params.beta.wireList <- rep(unique(params.beta.list),params.grid.length)
>
> temp <- stack(data.frame(z))
> negLog <- subset(temp, select=c(values))
> df.wireFrame <- data.frame(params.alpha.wireList,params.beta.wireList,negLog)
>
> # Plot parameter space
> wirePlot <- wireframe(values ~ params.alpha.wireList*params.beta.wireList, data=df.wireFrame, drape = TRUE,
+ col="purple",main="by Volkan OBAN using R - lattice",
+ col.regions = rainbow(100, s = 1, v = 1, start = 0, end = max(1,100-1)/100, alpha = 0.5),
+ xlab="Alpha", ylab="Beta", zlab="NLL",
+ screen = list (z = -140, x = -70, y = 3),
+ scales = list(arrows=FALSE,cex=.5,tick.number = 10))
> wirePlot
>

ggplot2
ref:http://rgraphgallery.blogspot.com.tr/2013/04/rg28-contour-plot.html
> require(ggplot2)
Zorunlu paket yükleniyor: ggplot2
> plt <- ggplot(gdr, aes(xvar, yvar, z= zvar)) + ggtitle("by Volkan OBAN using R - lattice \n contourplot - data:gdr ")
> plt + stat_contour() + theme_bw()
> plt + geom_tile(aes(fill = zvar)) + stat_contour() + theme_bw()
> plt + stat_contour(geom="polygon", aes(fill=..level..)) + theme_bw()
> require(ggplot2)
> plt <- ggplot(gdr, aes(xvar, yvar, z= zvar))
> plt + stat_contour() + theme_bw()
> plt + geom_tile(aes(fill = zvar)) + stat_contour() + theme_bw()
> plt + stat_contour(geom="polygon", aes(fill=..level..)) + theme_bw()
>

ggplot2
> require(ggplot2)
> plt <- ggplot(gdr, aes(xvar, yvar, z= zvar))
> plt + stat_contour() + theme_bw()

lattice package
> xvr <- seq(-5, 5, len = 50)
> yvr <- seq(-5, 5, len = 50)
> gdr <- expand.grid(xvar = xvr, yvar = yvr)
> gdr$zvar <- rnorm (nrow(gdr), 4, 1)
>
>
> #plot
> require(lattice)
> contourplot(zvar ~ xvar * yvar, data = gdr,main="by Volkan OBAN using R - lattice", cuts = 10)
> xvr <- seq(-5, 5, len = 50)
> yvr <- seq(-5, 5, len = 50)
> gdr <- expand.grid(xvar = xvr, yvar = yvr)
> gdr$zvar <- rnorm (nrow(gdr), 4, 1)

lattice package
> x <- seq(1,2,0.2);
> y <- seq(0.5,1.5,0.1);
>
> data1 <- matrix(0,nrow=length(x)*length(y),ncol=3);
> data2 <- matrix(0,nrow=length(x)*length(y),ncol=3);
>
> n <- 0;
> j <- 1;
> while(j<=length(x)){
+ for (k in 1:length(y)){
+ data1[k+n,1] <- x[j];
+ data1[k+n,2] <- y[k];
+ data1[k+n,3] <- x[j]^4 + y[k];
+
+ data2[k+n,1] <- x[j];
+ data2[k+n,2] <- y[k];
+ data2[k+n,3] <- x[j]^4 + y[k]^4 + 3;
+ }
+ n <- n+length(y);
+ j <- j+1;
+ }
> rm(x,y,j,n,k)
>
> # Arranging data into a data frame
> data1_2 <-as.data.frame(rbind(data1,data2));
> colnames(data1_2) <- c("x","y","z");
> data1_2$group <- gl(2, nrow(data1_2)/2, labels=c("data1", "data2"))
> rm(data1,data2)
>
> # Plotting data as a surface
> wireframe(z~x*y,data=data1_2,groups=group,
+
+ # Naming labels and Axis
+ main =list(label="by Volkan OBAN using R - lattice - wireframe ",cex=2,distance=5),
+ zlab=list(rot=90,label = "Z",cex=2),
+ xlab=list(label = "X",cex=2),
+ ylab=list(label = "Y",cex=2),
+
+ # Coloring the groups
+ col.groups=c(rgb(red=200,green=100,blue=80,
+ alpha=200,maxColorValue=255), # Orange
+ rgb(red=150,green=200,blue=205,
+ alpha=200,maxColorValue=255)), # Blue
+
+ # Coloring the grids
+ col=c(rgb(red=0,green=0,blue=0,alpha=50,maxColorValue=255),
+ rgb(red=0,green=0,blue=0,alpha=50,maxColorValue=255)),
+
+ aspect=c(1,1), # y-size/x-size and z-size/x-size
+ screen = list(z=40,y=0,x=-80)); # axis rotation
>

lattice package
> df <- data.frame(expand.grid(1:10,1:10),rep(10,100))
> colnames(df) <- c("x","y","z")
> wireframe(z~x*y,df,colorkey=TRUE,drape=TRUE)
>wireframe(z~x*y,df,main="",colorkey=TRUE,drape=TRUE, zlim=c(0,10))

lattice package
ref:http://zoonek.free.fr/blosxom/R/2006-08-10_R_Graphics.html
# Minimum Spanning Tree (MST)
panel.mst <- function (x, y, ...) {
require(ape) # For mst()
d <- dist(cbind(x,y))
m <- mst(d)
i <- which(m == 1)
panel.segments(x[row(m)[i]], y[row(m)[i]],
x[col(m)[i]], y[col(m)[i]],
...)
}
# 2-dimensional Kernel Density Estimation
panel.kde <- function (x, y, ...) {
require(grid) # for convertX() and unit()
require(MASS) # For kde2d()
k <- kde2d(
x, y,
n = 500,
# The limits of the current plot
lims = c(as.numeric(convertX(unit(0,"npc"),"native")),
as.numeric(convertX(unit(1,"npc"),"native")),
as.numeric(convertY(unit(0,"npc"),"native")),
as.numeric(convertY(unit(1,"npc"),"native"))))
panel.levelplot(rep(k$x, length(k$y)),
rep(k$y, each = length(k$x)),
sqrt(k$z),
subscripts = 1:length(k$z),
...)
}
# The same example as above
library(RColorBrewer)
xyplot(lat ~ long | Depth, data = quakes,
panel = function (x, y, ...) {
panel.kde(x, y,
col.regions = brewer.pal(9, "YlOrRd"))
panel.mst(x, y,
col = "black", lwd = 2)
},
strip = strip.custom(strip.names = TRUE,
strip.levels = TRUE),
par.strip.text = list(cex = 0.75),
aspect = "iso")

Plot3D package
X <- seq(0, pi, length.out = 50)
>
> Y <- seq(0, 2*pi, length.out = 50)
>
> M <- mesh(X, Y)
>
> phi <- M$x
>
> theta <- M$y
>
> # x, y and z grids
> x <- sin(phi) * cos(theta)
>
> y <- cos(phi)
>
> z <- sin(phi) * sin(theta)
>
> # these are the defaults
> p <- list(ambient = 0.3, diffuse = 0.6, specular = 1.,exponent = 20, sr = 0, alpha = 1)
>
> par(mfrow = c(3, 3), mar = c(0, 0, 0, 0))
>
> Col <- "magenta4"
>
> surf3D(x, y, z, box = FALSE, col = Col, lighting = TRUE)
> surf3D(x, y, z, box = FALSE, col = Col, lighting = list(exponent = 5))
> surf3D(x, y, z, box = FALSE, col = Col, lighting = list(exponent = 50))
> surf3D(x, y, z, box = FALSE, col = Col, shade = 0.9)
> surf3D(x, y, z, box = FALSE, col = Col, lighting = list(sr = 1))
> surf3D(x, y, z, box = FALSE, col = Col, lighting = list(diffuse = 0))
> surf3D(x, y, z, box = FALSE, col = Col, lighting = list(exponent = 50))
> surf3D(x, y, z, box = FALSE, col = Col, lighting = list(exponent = 20))
> surf3D(x, y, z, box = FALSE, col = Col, lighting = list(exponent = 1))
>

Plot3D package
image2D
Plot3D package
box3D(x0 = runif(4), y0 = runif(4), z0 = runif(4),
+ x1 = runif(4), y1 = runif(4), z1 = runif(4),
+ col = c("purple", "pink", "lightpink4"), alpha = 0.5,
+ border = "black", lwd = 2)

Plot3D package
z <- seq(0, 10, 0.2)
> x <- cos(z)
> y <- sin(z)*z
> scatter3D(x, y, z, phi = 0, bty = "g", type = "h", ticktype = "detailed")

Plot3D package
x <- y <- z <- seq(-1, 1, by = 0.1)
> grid <- mesh(x, y, z)
> colvar <- with(grid, x*exp(-x^2 - y^2 - z^2))
slice3D (x, y, z, colvar = colvar, theta = 60)
> slicecont3D (x, y, z, ys = seq(-1, 1, by = 0.5), colvar = colvar, theta = 60, border = "purple")
Plot3D package
a <- volcano[seq(1, 87, 15), seq(1, 61, 15)]
hist3D(z = a, scale = FALSE, expand = 0.01, bty = "g", phi = 20,
+ col = "#9932CC", border = "white", shade = 0.2, ltheta = 90, space = 0.3, ticktype = "detailed", d = 2)
Plot3D package
rect3D(x0 = seq(-0.8, -0.1, by = 0.1),
+ y0 = seq(-0.8, -0.1, by = 0.1),
+ z0 = seq(-0.8, -0.1, by = 0.1),
+ x1 = seq(0.8, 0.1, by = -0.1),
+ y1 = seq(0.8, 0.1, by = -0.1),
+ col = rainbow(8), border = "pink",
+ bty = "g", lwd = 2, phi = 20, main = " rect3D")
Plot3D package
box3D(x0 = seq(-0.8, -0.1, by = 0.1),
+ y0 = seq(-0.8, -0.1, by = 0.1),
+ z0 = seq(-0.8, -0.1, by = 0.1),
+ x1 = seq(0.8, 0.1, by = -0.1),
+ y1 = seq(0.8, 0.1, by = -0.1),
+ z1 = seq(0.8, 0.1, by = -0.1),
+ col = rainbow(n = 8, alpha = 0.1),
+ border = "purple", lwd = 2, phi = 20)
ref: https://rpubs.com/yoshio/95844
Plot3D package
> border3D(x0 = seq(-0.8, -0.1, by = 0.1),
+ y0 = seq(-0.8, -0.1, by = 0.1),
+ z0 = seq(-0.8, -0.1, by = 0.1),
+ x1 = seq(0.8, 0.1, by = -0.1),
+ y1 = seq(0.8, 0.1, by = -0.1),
+ z1 = seq(0.8, 0.1, by = -0.1),
+ col = rainbow(8), lty = 2,
+ lwd = c(1, 4), phi = 20, main = "")
Plot3D package
with (mtcars, {
# linear regression
fit <- lm(mpg ~ wt + disp)
# predict values on regular xy grid
wt.pred <- seq(1.5, 5.5, length.out = 30)
disp.pred <- seq(71, 472, length.out = 30)
xy <- expand.grid(wt = wt.pred,
disp = disp.pred)
mpg.pred <- matrix (nrow = 30, ncol = 30,
data = predict(fit, newdata = data.frame(xy),
interval = "prediction"))
# fitted points for droplines to surface
fitpoints <- predict(fit)
scatter3D(z = mpg, x = wt, y = disp, pch = 18, cex = 2,
theta = 20, phi = 20, ticktype = "detailed",
xlab = "wt", ylab = "disp", zlab = "mpg",
surf = list(x = wt.pred, y = disp.pred, z = mpg.pred,
facets = NA, fit = fitpoints),
main = "")
})
ref:https://rpubs.com/yoshio/95844

Plot3D package
reference:https://rpubs.com/yoshio/95844
> X <- seq(0, pi, length.out = 50)
> Y <- seq(0, 2*pi, length.out = 50)
> M <- mesh(X, Y)
> phi <- M$x
> theta <- M$y
> r <- sin(4*phi)^3 + cos(2*phi)^3 + sin(6*theta)^2 + cos(6*theta)^4
> x <- r * sin(phi) * cos(theta)
> y <- r * cos(phi)
> z <- r * sin(phi) * sin(theta)
> surf3D(x, y, z, colvar = y, colkey = FALSE, shade = 0.5,box = FALSE, theta = 60)
> surf3D(x, y, z, colvar = y, colkey = FALSE, box = FALSE, theta = 60, facets = FALSE

Plot3D package
> x <- rchisq(1000, df = 5)
> hs <- hist(x, breaks = 20)
hist3D(x = hs$mids, y = 1, z = matrix(ncol = 1, data = hs$density), bty = "g", ylim = c(0., 2.0), scale = FALSE, expand = 20, border = "pink", col = "red", shade = 0.4, space = 0.1, theta = 20, phi = 20, main = "")
Plot3D package
volkan <- volcano[seq(1, 87, 15), seq(1, 61, 15)]
ribbon3D(z = volkan, scale = FALSE, expand = 0.01, bty = "g", phi = 20, col = "pink", border = "purple", shade = 0.2, ltheta = 90,space = 0.3, ticktype = "detailed", d = 2, curtain = TRUE)

Plot3D package
hist3D
Plot3D package
> x <- y <- z <- seq(-4, 4, by = 0.2)
> M <- mesh(x, y, z)
> R <- with (M, sqrt(x^2 + y^2 + z^2))
> p <- sin(2*R) /(R+1e-3)
> slice3D(x, y, z, colvar = p, d = 2, theta = 60, border = "black", xs = c(-4, 0), ys = c(-4, 0, 4), zs = c(-4, 0))

geofacet
library(ggplot2)
library(geofacet)
ggplot(eu_imm, aes(year, persons)) +
+ geom_line() +
+ facet_geo(~ name, grid = "eu_grid1") +
+ scale_x_continuous(labels = function(x) paste0("'", substr(x, 3, 4))) +
+ scale_y_sqrt(minor_breaks = NULL) +
+ ylab("# Resettled Persons") +
+ theme_bw()

time series forecasting
# Load packages
library(forecast) # Most popular forecasting pkg
library(sweep) # Broom tidiers for forecast pkg
library(timekit) # Working with time series in R
library(tidyquant) # Get's data from FRED, loads tidyverse behind the scenes
library(geofacet)
> ne_gdp <- tq_get("NENGSP", get = "economic.data", from = "2007-01-01", to = "2017-06-01") %>%
+ rename(gdp = price)
> states <- tibble(abbreviation = state.abb) %>%
+ mutate(fred_code = paste0(abbreviation, "NGSP")) %>%
+ select(2:1)
> states_gdp <- states %>%
+ tq_get(get = "economic.data", from = "2007-01-01", to = "2017-06-01")
>
> # Group and rename
> states_gdp <- states_gdp %>%
+ select(-fred_code) %>%
+ group_by(abbreviation) %>%
+ rename(gdp = price)
> ne_gdp_ts <- ne_gdp %>%
+ tk_ts(start = 2017, freq = 1, silent = TRUE)
> ne_fit_arima <- auto.arima(ne_gdp_ts)
> sw_glance(ne_fit_arima)
# A tibble: 1 x 12
model.desc sigma logLik AIC BIC
<chr> <dbl> <dbl> <dbl> <dbl>
1 ARIMA(0,1,0) with drift 2149.529 -81.29672 166.5934 166.9879
# ... with 7 more variables: ME <dbl>, RMSE <dbl>, MAE <dbl>,
# MPE <dbl>, MAPE <dbl>, MASE <dbl>, ACF1 <dbl>
> ne_fcast <- forecast(ne_fit_arima, h = 3)
> ne_sweep <- sw_sweep(ne_fcast, timekit_idx = TRUE, rename_index = "date")
> ne_sweep
# A tibble: 13 x 7
date key gdp lo.80 lo.95 hi.80 hi.95
<date> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 2007-01-01 actual 81926.0 NA NA NA NA
2 2008-01-01 actual 84873.0 NA NA NA NA
3 2009-01-01 actual 86961.0 NA NA NA NA
4 2010-01-01 actual 92231.0 NA NA NA NA
5 2011-01-01 actual 99935.0 NA NA NA NA
6 2012-01-01 actual 101973.0 NA NA NA NA
7 2013-01-01 actual 106765.0 NA NA NA NA
8 2014-01-01 actual 112087.0 NA NA NA NA
9 2015-01-01 actual 113458.0 NA NA NA NA
10 2016-01-01 actual 115345.0 NA NA NA NA
11 2017-01-01 forecast 119058.2 116303.5 114845.2 121813.0 123271.2
12 2018-01-01 forecast 122771.4 118875.7 116813.4 126667.2 128729.5
13 2019-01-01 forecast 126484.7 121713.3 119187.5 131256.0 133781.8
> ne_sweep %>%
+ ggplot(aes(x = date, y = gdp, color = key)) +
+ # Prediction intervals
+ geom_ribbon(aes(ymin = lo.95, ymax = hi.95),
+ fill = "#D5DBFF", color = NA, size = 0) +
+ geom_ribbon(aes(ymin = lo.80, ymax = hi.80, fill = key),
+ fill = "#596DD5", color = NA, size = 0, alpha = 0.8) +
+ # Actual & Forecast
+ geom_line(size = 1) +
+ geom_point(size = 2) +
+ # Aesthetics
+ theme_tq(base_size = 16) +
+ scale_color_tq() +
+ labs(title = " by Volkan OBAN using R : forecast-sweep-geofacet-timelit-tidyquant packages \n Nebraska GDP, 3-Year Forecast", x = "", y = "GDP, USD Millions")
> states_gdp <- states_gdp %>%
+ nest()
> states_gdp
# A tibble: 50 x 2
abbreviation data
<chr> <list>
1 AL <tibble [10 x 2]>
2 AK <tibble [10 x 2]>
3 AZ <tibble [10 x 2]>
4 AR <tibble [10 x 2]>
5 CA <tibble [10 x 2]>
6 CO <tibble [10 x 2]>
7 CT <tibble [10 x 2]>
8 DE <tibble [10 x 2]>
9 FL <tibble [10 x 2]>
10 GA <tibble [10 x 2]>
# ... with 40 more rows
> states_gdp <- states_gdp %>%
+ mutate(data_ts = map(data, tk_ts, freq = 1, start = 2007, silent = TRUE))
> states_gdp
# A tibble: 50 x 3
abbreviation data data_ts
<chr> <list> <list>
1 AL <tibble [10 x 2]> <S3: ts>
2 AK <tibble [10 x 2]> <S3: ts>
3 AZ <tibble [10 x 2]> <S3: ts>
4 AR <tibble [10 x 2]> <S3: ts>
5 CA <tibble [10 x 2]> <S3: ts>
6 CO <tibble [10 x 2]> <S3: ts>
7 CT <tibble [10 x 2]> <S3: ts>
8 DE <tibble [10 x 2]> <S3: ts>
9 FL <tibble [10 x 2]> <S3: ts>
10 GA <tibble [10 x 2]> <S3: ts>
# ... with 40 more rows
> states_gdp <- states_gdp %>%
+ mutate(fit = map(data_ts, auto.arima))
> states_gdp
# A tibble: 50 x 4
abbreviation data data_ts fit
<chr> <list> <list> <list>
1 AL <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
2 AK <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
3 AZ <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
4 AR <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
5 CA <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
6 CO <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
7 CT <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
8 DE <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
9 FL <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
10 GA <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
# ... with 40 more rows
> states_gdp %>%
+ mutate(glance = map(fit, sw_glance)) %>%
+ unnest(glance, .drop = T)
# A tibble: 50 x 13
abbreviation model.desc sigma logLik
<chr> <chr> <dbl> <dbl>
1 AL ARIMA(0,1,0) with drift 3267.828 -85.06590
2 AK ARIMA(0,0,0) with non-zero mean 4199.313 -97.08934
3 AZ ARIMA(0,2,0) 7559.654 -82.79488
4 AR ARIMA(0,1,0) with drift 2231.839 -81.63464
5 CA ARIMA(0,2,0) 60035.965 -99.37208
6 CO ARIMA(0,1,0) with drift 7064.218 -92.00497
7 CT ARIMA(0,2,0) 5009.932 -79.50274
8 DE ARIMA(0,1,0) with drift 1865.871 -80.02328
9 FL ARIMA(0,2,0) 17001.163 -89.27758
10 GA ARIMA(0,2,0) 6369.686 -81.42147
# ... with 40 more rows, and 9 more variables: AIC <dbl>,
# BIC <dbl>, ME <dbl>, RMSE <dbl>, MAE <dbl>, MPE <dbl>,
# MAPE <dbl>, MASE <dbl>, ACF1 <dbl>
> states_gdp <- states_gdp %>%
+ mutate(forecast = map(fit, forecast, h = 3))
> states_gdp
# A tibble: 50 x 5
abbreviation data data_ts fit
<chr> <list> <list> <list>
1 AL <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
2 AK <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
3 AZ <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
4 AR <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
5 CA <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
6 CO <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
7 CT <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
8 DE <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
9 FL <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
10 GA <tibble [10 x 2]> <S3: ts> <S3: ARIMA>
# ... with 40 more rows, and 1 more variables: forecast <list>
> states_gdp_sweep <- states_gdp %>%
+ mutate(sweep = map(forecast, sw_sweep, timekit_idx = T, rename_index = "date")) %>%
+ select(abbreviation, sweep) %>%
+ unnest()
> states_gdp_sweep
# A tibble: 650 x 8
abbreviation date key gdp lo.80 lo.95 hi.80 hi.95
<chr> <date> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 AL 2007-01-01 actual 169923 NA NA NA NA
2 AL 2008-01-01 actual 172646 NA NA NA NA
3 AL 2009-01-01 actual 168315 NA NA NA NA
4 AL 2010-01-01 actual 174710 NA NA NA NA
5 AL 2011-01-01 actual 180665 NA NA NA NA
6 AL 2012-01-01 actual 185878 NA NA NA NA
7 AL 2013-01-01 actual 190319 NA NA NA NA
8 AL 2014-01-01 actual 194404 NA NA NA NA
9 AL 2015-01-01 actual 199980 NA NA NA NA
10 AL 2016-01-01 actual 204861 NA NA NA NA
# ... with 640 more rows
> states_gdp_sweep %>%
+ ggplot(aes(x = date, y = gdp, color = key)) +
+ # Prediction intervals
+ geom_ribbon(aes(ymin = lo.95, ymax = hi.95),
+ fill = "#D5DBFF", color = NA, size = 0) +
+ geom_ribbon(aes(ymin = lo.80, ymax = hi.80, fill = key),
+ fill = "#596DD5", color = NA, size = 0, alpha = 0.8) +
+ # Actual & Forecast
+ geom_line() +
+ # Aesthetics
+ scale_y_continuous(label = function(x) x*1e-6) +
+ scale_x_date(date_breaks = "5 years", labels = scales::date_format("%Y")) +
+ facet_geo(~ abbreviation, scale = "free_y") +
+ theme_tq() +
+ scale_color_tq() +
+ theme(legend.position = "none",
+ axis.text.x = element_text(angle = 45, hjust = 1),
+ axis.text.y = element_blank()
+ ) +
+ ggtitle(" State GDP, 3-Year Forecast") +
+ xlab("") +
+ ylab("GDP, Free Scale")
> states_gdp_sweep %>%
+ ggplot(aes(x = date, y = gdp, color = key)) +
+ # Prediction intervals
+ geom_ribbon(aes(ymin = lo.95, ymax = hi.95),
+ fill = "#D5DBFF", color = NA, size = 0) +
+ geom_ribbon(aes(ymin = lo.80, ymax = hi.80, fill = key),
+ fill = "#596DD5", color = NA, size = 0, alpha = 0.8) +
+ # Actual & Forecast
+ geom_line() +
+ # Aesthetics
+ scale_y_continuous(label = function(x) x*1e-6) +
+ scale_x_date(date_breaks = "5 years", labels = scales::date_format("%Y")) +
+ facet_geo(~ abbreviation, scale = "free_y") +
+ theme_tq() +
+ scale_color_tq() +
+ theme(legend.position = "none",
+ axis.text.x = element_text(angle = 45, hjust = 1),
+ axis.text.y = element_blank()
+ ) +
+ ggtitle(" by Volkan OBAN using R :TIME SERIES FORECASTING - forecast-sweep-geofacet-timelit-tidyquant packages \n State GDP, 3-Year Forecast") +
+ xlab("") +
+ ylab("GDP, Free Scale")
> states_gdp_sweep %>%
+ ggplot(aes(x = date, y = gdp, color = key)) +
+ # Prediction intervals
+ geom_ribbon(aes(ymin = lo.95, ymax = hi.95),
+ fill = "#D5DBFF", color = NA, size = 0) +
+ geom_ribbon(aes(ymin = lo.80, ymax = hi.80, fill = key),
+ fill = "#596DD5", color = NA, size = 0, alpha = 0.8) +
+ # Actual & Forecast
+ geom_line() +
+ # Aesthetics
+ scale_y_continuous(label = function(x) x*1e-6) +
+ scale_x_date(date_breaks = "5 years", labels = scales::date_format("%Y")) +
+ facet_geo(~ abbreviation, scale = "free_y") +
+ theme_tq() +
+ scale_color_tq() +
+ theme(legend.position = "none",
+ axis.text.x = element_text(angle = 45, hjust = 1),
+ axis.text.y = element_blank()
+ ) +
+ ggtitle(" State GDP, 3-Year Forecast") +
+ xlab("") +
+ ylab("GDP, Free Scale")
> states_gdp_sweep %>%
+ ggplot(aes(x = date, y = gdp, color = key)) +
+ # Prediction intervals
+ geom_ribbon(aes(ymin = lo.95, ymax = hi.95),
+ fill = "#D5DBFF", color = NA, size = 0) +
+ geom_ribbon(aes(ymin = lo.80, ymax = hi.80, fill = key),
+ fill = "#596DD5", color = NA, size = 0, alpha = 0.8) +
+ # Actual & Forecast
+ geom_line() +
+ # Aesthetics
+ scale_y_continuous(label = function(x) x*1e-6) +
+ scale_x_date(date_breaks = "5 years", labels = scales::date_format("%Y")) +
+ facet_geo(~ abbreviation, scale = "free_y") +
+ theme_tq() +
+ scale_color_tq() +
+ theme(legend.position = "none",
+ axis.text.x = element_text(angle = 45, hjust = 1),
+ axis.text.y = element_blank()
+ ) +
+ ggtitle(" State GDP, 3-Year Forecast") +
+ xlab("") +
+ ylab("GDP, Free Scale")

rms package
plot.xmean.ordinaly

rms package - nomogram
w <- upData(d,
cens = 15 * runif(n),
h = .02 * exp(.04 * (age - 50) + .8 * (sex == 'Female')),
d.time = -log(runif(n)) / h,
death = ifelse(d.time <= cens, 1, 0),
d.time = pmin(d.time, cens))
f <- psm(Surv(d.time,death) ~ sex * age, data=w, dist='lognormal')
med <- Quantile(f)
surv <- Survival(f) # This would also work if f was from cph
plot(nomogram(f, fun=function(x) med(lp=x), funlabel="Median Survival Time"))

rms package
> n <- 1000 # define sample size
> set.seed(17) # so can reproduce the results
> age <- rnorm(n, 50, 10)
> blood.pressure <- rnorm(n, 120, 15)
> cholesterol <- rnorm(n, 200, 25)
> sex <- factor(sample(c('female','male'), n,TRUE))
> label(age) <- 'Age' # label is in Hmisc
> label(cholesterol) <- 'Total Cholesterol'
> label(blood.pressure) <- 'Systolic Blood Pressure'
> label(sex) <- 'Sex'
> units(cholesterol) <- 'mg/dl' # uses units.default in Hmisc
> units(blood.pressure) <- 'mmHg'
> # Specify population model for log odds that Y=1
> L <- .4*(sex=='male') + .045*(age-50) +
+ (log(cholesterol - 10)-5.2)*(-2*(sex=='female') + 2*(sex=='male'))
> # Simulate binary y to have Prob(y=1) = 1/[1+exp(-L)]
> y <- ifelse(runif(n) < plogis(L), 1, 0)
> ddist <- datadist(age, blood.pressure, cholesterol, sex)
> options(datadist='ddist')
> fit <- lrm(y ~ blood.pressure + sex * (age + rcs(cholesterol,4)),
+ x=TRUE, y=TRUE)
> p <- Predict(fit, age, cholesterol, sex, np=50) # vary sex last
> bplot(p, main="by Volkan OBAN using R - rms package")
> bplot(p,, main="by Volkan OBAN using R - rms package", lfun=wireframe)

quandl package
plot(stl(Quandl("WIKI/GOOG",type="ts",collapse="monthly")[,11],s.window="per"))
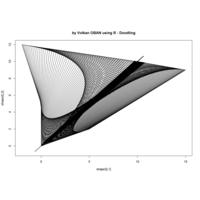
Doodling
doodle <- function(
start=c(0,0),
targets = rbind(c(0,10),c(10,10), c(10,0), c(0,0)) ,
tdist = .25,
speed = c(0,0),
accel = .1,
resis = .005,
jitter = .0005,
chncStp = 0) {
# start - We start with the starting position
# targ - Points that will be pursued (initially just a square)
# tdist - How close we need to get to each point before moving on
# speed - Initial speed
# accel - How fast does the drawer accelerate towards that point
# resis - What percentage of speed is lost each round
# jitter - A normal draw random jitter that moves the writing tool in an unexpected direction.
# chncStp - There is some chance that the drawing tool will kill all momentum and stop.
# First off I define a function uvect to convert any two sets of points
# into a unit vector and measure the distance between the two points.
uvect <- function(p1,p2=NULL) {
if (is.null(p2)) {
p2 <- p1[[2]]
p1 <- p1[[1]]
}
list(vect=(p2-p1)/sqrt(sum((p1-p2)^2)), dist=sqrt(sum((p1-p2)^2)))
}
# Starup parameters
i <- 1
plist <- position <- start # plist saves all of the points that the drawing tool has passed through
vect <- uvect(position,targets[i,])
while(i<=nrow(targets)) {
# Calculate the appropriate unit vector and distance from end point
vect <- uvect(position,targets[i,])
# Remove some amount of speed from previous velocity
speed <- speed*(1-resis)
# IF drawer randomly stops remove all speed
if (rbinom(1,1,chncStp)) speed<-0
#
speed <- speed + accel*vect[[1]] + rnorm(2)*jitter
position <- position + speed
plist <- rbind(plist,position)
vect <- uvect(position,targets[i,])
if (vect[[2]]<tdist) i <- i+1
}
plist
}
plist <- doodle()
shape <- doodle(cbind(c(0,-2,10,15,5,0),c(5,12,10,9,2,0)),resis=.2)
>
> plot(shape, type="l",lwd=1)
>
tidygraph
plot(play_forestfire(40, 0.8))
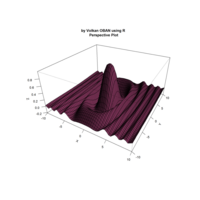
persp-- Perspective Plots
> x.coord <- seq(-10, 10, length= 50)
> y.coord <- x.coord
> func <- function(x,y) { r <- sqrt(abs(x^3)+y^2); sin(r)/r }
> z.coord <- outer(x.coord, y.coord, func)
> persp(x.coord,y.coord,z.coord,theta=30,phi=30,expand=0.5,col="hotpink4",
+ ltheta=120,shade=0.75,ticktype="detailed",xlab="X",ylab="Y",zlab="Z")
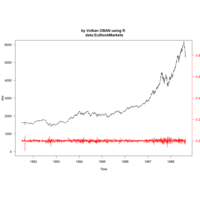
Plot
> data("EuStockMarkets")
> dax <- EuStockMarkets[, 1]
> plot(dax, ylim = c(0, 6000), axes = FALSE)
> axis(1)
> axis(2, las = 1)
> par(new = TRUE)
> plot(diff(log(dax)), ylim = c(-0.1, 0.9), axes = FALSE, col = 2, ylab = "")
> box()
> axis(4, col = 2, col.axis = 2, las = 1)

persp-- Perspective Plots
> y <- x <- seq(-3,3,length=50)
> f <- function(x,y){
+ dnorm(x^4)*dnorm(y^2)}
> z <- outer(x,y,f)
> persp(x,y,z, theta = 60, phi = 30,col = "lightpink1",zlim=c(0,0.2))
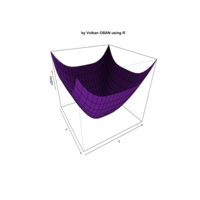
persp-- Perspective Plots
> cone <- function(x, y){
+ sqrt(x^4+y^4)
+ }
> x <- y <- seq(-1, 1, length= 20)
> z <- outer(x, y, cone)
> persp(x, y, z)

treemap-- d3treeR
library(treemap)
library(d3treeR)
# dataset
group=c(rep("group-1",4),rep("group-2",2),rep("group-3",3))
subgroup=paste("subgroup" , c(1,2,3,4,1,2,1,2,3), sep="-")
value=c(13,5,22,12,11,7,3,1,23)
data=data.frame(group,subgroup,value)
# basic treemap
p=treemap(data,
index=c("group","subgroup"),
vSize="value",
type="index"
)
# make it interactive ("rootname" becomes the title of the plot):
inter=d3tree2( p , rootname = "General" )

cartography package
library(cartography)
# Upload data attached with the package.
data(nuts2006)
# Now we have a spdf file (shape file) called nuts2.spdf with shape of european regions. We can plot it with the plot function.
summary(nuts2.spdf)
# We also have a dataframe with information concerning every region.
head(nuts2.df)
# Both object have a first column "id" that makes the link between them.
# Create a grid
mygrid <- getGridLayer(spdf = nuts2.spdf, cellsize = 2e+05)
# You can plot the grid
# plot(mygrid$spdf)
# Adapt grid to a numerical variable
datagrid.df <- getGridData(x = mygrid, df = nuts2.df, var = "pop2008")
datagrid.df$densitykm <- datagrid.df$pop2008_density * 1000 * 1000
# Plot background
plot(nuts0.spdf, border = NA, col = NA, bg = "#A6CAE0")
plot(world.spdf, col = "#E3DEBF", border = NA, add = TRUE)
# Plot density of population
choroLayer(spdf = mygrid$spdf, df = datagrid.df, var = "densitykm",
border = "grey80", col = carto.pal(pal1 = "wine.pal", n1 = 6),
legend.pos = "topright", method = "q6",
add = TRUE, legend.title.txt = "Population Density\n(inhabitant/km²)")
# Title, legend..
layoutLayer(title = "Population Density", coltitle = "black",
col = NA, sources = "Eurostat, 2011", scale = NULL,
author = "cartography", frame = FALSE)

igraph
g <- barabasi.game(5000, power=1)
> layout <- layout.fruchterman.reingold(g)
> membership <- cut_at(eb, no = 10)
> plot(g,
+ vertex.color= rainbow(10, .8, .8, alpha=.8)[membership],
+ vertex.size=5, layout=layout, vertex.label=NA,
+ edge.arrow.size=.2)
> eb <- walktrap.community(g)
> membership <- cut_at(eb, no = 10)
> plot(g,
+ vertex.color= rainbow(10, .8, .8, alpha=.8)[membership],
+ vertex.size=5, layout=layout, vertex.label=NA,
+ edge.arrow.size=.2)

igraph
g <- barabasi.game(10000, power=1)
> layout <- layout.fruchterman.reingold(g)
> plot(g, layout=layout, vertex.size=2, vertex.label=NA, edge.arrow.size=.2)

wireframe
wireframe(z ~ x * y, data = g, groups = gr,
scales = list(arrows = FALSE,
x = list(at = c(2, 5, 10)),
y = list(at = c(6, 10, 14),
lab = c('A', 'BBB', 'CCCCC'))
))
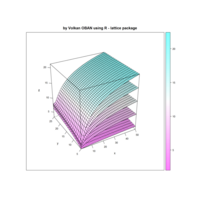
lattice package
> g <- expand.grid(x = 1:50, y = 5:25, gr = 1:5)
> g$z <- log((g$x^g$gr + g$y^2) * g$gr)
> wireframe(z ~ x * y, data = g, groups = gr,
+ scales = list(arrows = FALSE),
+ drape = TRUE, colorkey = TRUE,main="by Volkan OBAN using R - lattice package",
+ screen = list(z = 30, x = -60))
Plot
> U = numeric(1000);
> n = 100;
> average = numeric(n);
> for (i in 1 : n)
+ {U = runif(1000);
+ X = tan(pi ∗ (U − 0.5));
+ average[i] = mean(X); }
> plot(1 : n, average[1 : n], type = "l", lwd = 2, col = "red",main="by Volkan OBAN using R") + theme_solarized(light = FALSE) +
+ scale_colour_solarized("red")

mandelbrot package
> par(mfrow = c(1, 2), pty = "s", mar = rep(0, 4))
> plot(mb,col = cols, transform = "inverse")
> plot(mb, col = cols, transform = "log")
ref:https://github.com/blmoore/

mandelbrot package
> library(ggplot2)
>
> mb <- mandelbrot(xlim = c(-0.8335, -0.8325),
+ ylim = c(0.205, 0.206),
+ resolution = 1200L,
+ iterations = 1000)
>
>
> cols <- c(
+ colorRampPalette(c("#e7f0fa", "#c9e2f6", "#95cbee",
+ "#0099dc", "#4ab04a", "#ffd73e"))(10),
+ colorRampPalette(c("#eec73a", "#e29421", "#e29421",
+ "#f05336","#ce472e"), bias=2)(90),
+ "black")
>
> df <- as.data.frame(mb)
> ggplot(df, aes(x = x, y = y, fill = value)) +
+ geom_raster(interpolate = TRUE) + theme_void() +
+ scale_fill_gradientn(colours = cols, guide = "none") + ggtitle("by Volkan OBAN using R-mandelbrot package ")
> library(ggplot2)
>
> mb <- mandelbrot(xlim = c(-0.8335, -0.8325),
+ ylim = c(0.205, 0.206),
+ resolution = 1200L,
+ iterations = 1000)
>
>
> cols <- c(
+ colorRampPalette(c("#e7f0fa", "#c9e2f6", "#95cbee",
+ "#0099dc", "#4ab04a", "#ffd73e"))(10),
+ colorRampPalette(c("#eec73a", "#e29421", "#e29421",
+ "#f05336","#ce472e"), bias=2)(90),
+ "black")
>
> df <- as.data.frame(mb)
> ggplot(df, aes(x = x, y = y, fill = value)) +
+ geom_raster(interpolate = TRUE) + theme_void() +
+ scale_fill_gradientn(colours = cols, guide = "none")

sde package
t <- 0:100 # time
> sig2 <- 0.01
> nsim <- 1000
> ## we'll simulate the steps from a uniform distribution with limits set to
> ## have the same variance (0.01) as before
> X <- matrix(runif(n = nsim * (length(t) - 1), min = -sqrt(3 * sig2), max = sqrt(3 *
+ sig2)), nsim, length(t) - 1)
> X <- cbind(rep(0, nsim), t(apply(X, 1, cumsum)))
> plot(t, X[1, ], xlab = "time",ylab = "phenotype", ylim = c(-2, 2), type = "l")
> apply(X[2:nsim, ], 1, function(x, t) lines(t, x), t = t)
ggspectra pckg
library(photobiology)
plot(sun.spct) + theme_solarized(light = FALSE) +
+ scale_colour_solarized("red")
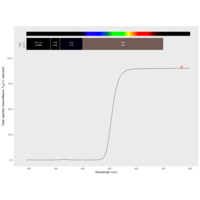
ggspectra pckg
library(photobiology)
plot(yellow_gel.spct)
plot(yellow_gel.spct, pc.out = TRUE)
ggraph ggthemes
graph <- graph_from_data_frame(flare$edges, vertices = flare$vertices)
set.seed(1)
ggraph(graph, 'circlepack', weight = 'size') +
geom_node_circle(aes(fill = depth), size = 0.25, n = 50) +
coord_fixed()
> ggraph(graph, 'circlepack', weight = 'size') +
+ geom_node_circle(aes(fill = depth), size = 0.25, n = 50) +
+ coord_fixed() + ggtitle("by Volkan OBAN using R-ggraph ") + theme_economist() + scale_colour_economist() +
+ scale_y_continuous(position = "right")

survminer package
ggsurvplot(
+ fit, # survfit object with calculated statistics.
+ data = lung, # data used to fit survival curves.
+ risk.table = TRUE, # show risk table.
+ pval = TRUE, # show p-value of log-rank test.
+ conf.int = TRUE, # show confidence intervals for
+ # point estimates of survival curves.
+ xlim = c(0,500), # present narrower X axis, but not affect
+ # survival estimates.
+ xlab = "Time in days", # customize X axis label.
+ break.time.by = 100, # break X axis in time intervals by 500.
+ ggtheme = theme_light(), # customize plot and risk table with a theme.
+ risk.table.y.text.col = T, # colour risk table text annotations.
+ risk.table.y.text = FALSE ,title="by Volkan OBAN using R - survminer"
+ )
>

ggTimeSeries
calenda HeatMap

ggmosaic package
ggplot(data = happy) +
+ geom_mosaic(aes(weight = wtssall, x = product(health), fill = health)) +
+ facet_grid(happy~.)
ggmosaic package
ggplot(data = happy) +
+ geom_mosaic(aes(weight=wtssall, x=product(health, sex, degree), fill=happy), na.rm=TRUE)
ggplot2 maps ggthemes
wm <- map("world",fill=TRUE,col=0,xlim=c(-10,40),ylim=c(30,60))
ggplot(wm, aes(long, lat, group = group)) +
+ geom_polygon(fill = "white", colour = "purple") + theme_economist() + scale_colour_economist() +
+ scale_y_continuous(position = "right")

ggplot2 ggalt ggthemes
> library(dplyr)
> library(tidyr)
> library(scales)
> library(ggplot2)
> library(ggalt) # devtools::install_github("hrbrmstr/ggalt")
>
> health <- read.csv("https://rud.is/dl/zhealth.csv", stringsAsFactors=FALSE,
+ header=FALSE, col.names=c("pct", "area_id"))
>
> areas <- read.csv("https://rud.is/dl/zarea_trans.csv", stringsAsFactors=FALSE, header=TRUE)
>
> health %>%
+ mutate(area_id=trunc(area_id)) %>%
+ arrange(area_id, pct) %>%
+ mutate(year=rep(c("2014", "2013"), 26),
+ pct=pct/100) %>%
+ left_join(areas, "area_id") %>%
+ mutate(area_name=factor(area_name, levels=unique(area_name))) -> health
>
> setNames(bind_cols(filter(health, year==2014), filter(health, year==2013))[,c(4,1,5)],
+ c("area_name", "pct_2014", "pct_2013")) -> health
>
> gg <- ggplot(health, aes(x=pct_2014, xend=pct_2013, y=area_name, group=area_name)) + ggtitle("by Volkan OBAN using R ")
> gg <- gg + geom_dumbbell(colour="#a3c4dc", size=1.5, colour_xend="#0e668b",
+ dot_guide=TRUE, dot_guide_size=0.15)
>
> gg
> gg + theme_wsj() + scale_colour_wsj("colors6", "")
> gg + theme_hc(bgcolor = "darkunica") +
+ scale_colour_hc("darkunica")
>

ggstance package
> library("ggstance")
>
> # Horizontal with ggstance
> ggplot(mpg, aes(hwy, class, fill = factor(cyl))) +
+ geom_boxploth()

ggplot2 and ggtech
ref:
http://kbroman.org/datacarpentry_R_2016-06-01/04-ggplot2.html
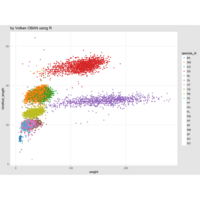
ggplot2 and ggthemes
ref:
http://kbroman.org/datacarpentry_R_2016-06-01/04-ggplot2.html

ggplot2 and ggthemes
ref:
http://kbroman.org/datacarpentry_R_2016-06-01/04-ggplot2.html
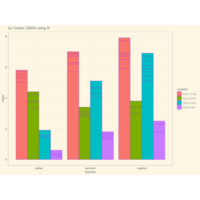
ggplot2 and ggthe
ref:
http://kbroman.org/datacarpentry_R_2016-06-01/04-ggplot2.html
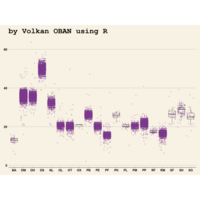
ggplot2
ref:
http://kbroman.org/datacarpentry_R_2016-06-01/04-ggplot2.html
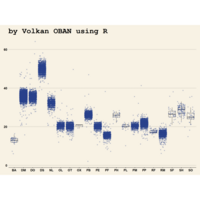
ggplot2 and ggthemes
ref:
http://kbroman.org/datacarpentry_R_2016-06-01/04-ggplot2.html

ggplot2
ref: http://kbroman.org/datacarpentry_R_2016-06-01/04-ggplot2.html
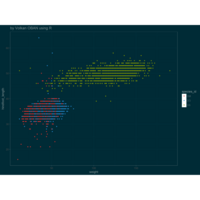
ggplot2
ref http://kbroman.org/datacarpentry_R_2016-06-01/04-ggplot2.html
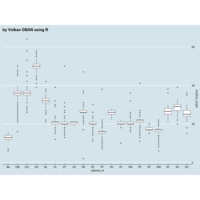
ggplot2 and ggthemes
a<- ggplot(surveys_complete, aes(x = species_id, y = hindfoot_length)) + geom_boxplot()
a + theme_economist() + scale_colour_economist() +
+ scale_y_continuous(position = "right"

epanetReader package-- plotSparklineTable
> plotSparklineTable(Theoph, row.var = 'Subject', col.vars = 'conc')

epanetReader package-- plotSparklineTable
> msr <- file.path( find.package("epanetReader"), "extdata","example.rpt")
> #read the results into R
> x <- read.msxrpt(msr)
> names(x)
[1] "Title" "nodeResults" "linkResults"
> summary(x)
plot(x)

sjplot--sjp.glm: plot probability curves (relationship between predictors and response)
> mydf <- data.frame(y = as.factor(y),
+ sex = to_factor(efc$c161sex),
+ dep = to_factor(efc$e42dep),
+ barthel = efc$barthtot,
+ education = to_factor(efc$c172code))
> # fit model
> fit <- glm(y ~., data = mydf, family = binomial(link = "logit"))
# plot probability curves (relationship between predictors and response)
> sjp.glm(fit, title = " Negative impact with 7 items", type = "slope")
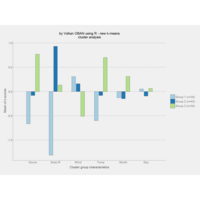
sjPlot and sjmisc package
airgrp <- sjc.qclus(airquality)
sjc.qclus(airquality, groupcount = 3, groups = airgrp$classification, title=" new k-means cluster analysis")
ggplot2
a<-ggplot(mtcars, aes(x = mpg^2, y = wt/cyl)) + geom_smooth(fill="purple",color="pink",size=2) + geom_jitter(color="darkgreen",shape=2) + geom_point(color="yellow") + ggtitle("by Volkan OBAN using R ")
a

Plot
> day=as.Date("2017-06-14") - 0:364
> value=runif(365) + seq(-140, 224)^2 / 10000
> data=data.frame(day, value)
> data %>% mutate(month = as.Date(cut(day, breaks = "month"))) %>%
+ ggplot(aes(x=day, y=value, fill=as.factor(month))) +
+ geom_line() +
+ geom_area() +
+ theme(
+ legend.position="none",
+ axis.text.x=element_blank(),
+ axis.ticks.x=element_blank(),
+ strip.background = element_rect(fill=alpha("slateblue",0.2)),
+ strip.placement="bottom"
+ ) +
+ xlab("by Volkan OBAN using R \n faceting for time series") +
+ facet_wrap(~as.Date(month), scales="free", ncol=3) + theme_tufte(ticks=FALSE) +
+ geom_tufteboxplot(median.type = "line", whisker.type = 'line', hoffset = 0, width = 3)

quantmod and plotly
library(plotly)
library(quantmod)
# get data
getSymbols("AAPL",src='yahoo')
df <- data.frame(Date=index(AAPL),coredata(AAPL))
# create Bollinger Bands
bbands <- BBands(AAPL[,c("AAPL.High","AAPL.Low","AAPL.Close")])
# join and subset data
df <- subset(cbind(df, data.frame(bbands[,1:3])), Date >= "2015-02-14")
# colors column for increasing and decreasing
for (i in 1:length(df[,1])) {
if (df$AAPL.Close[i] >= df$AAPL.Open[i]) {
df$direction[i] = 'Increasing'
} else {
df$direction[i] = 'Decreasing'
}
}
i <- list(line = list(color = '#17BECF'))
d <- list(line = list(color = '#7F7F7F'))
# plot candlestick chart
p <- df %>%
plot_ly(x = ~Date, type="candlestick",
open = ~AAPL.Open, close = ~AAPL.Close,
high = ~AAPL.High, low = ~AAPL.Low, name = "AAPL",
increasing = i, decreasing = d) %>%
add_lines(y = ~up , name = "B Bands",
line = list(color = '#ccc', width = 0.5),
legendgroup = "Bollinger Bands",
hoverinfo = "none") %>%
add_lines(y = ~dn, name = "B Bands",
line = list(color = '#ccc', width = 0.5),
legendgroup = "Bollinger Bands",
showlegend = FALSE, hoverinfo = "none") %>%
add_lines(y = ~mavg, name = "Mv Avg",
line = list(color = '#E377C2', width = 0.5),
hoverinfo = "none") %>%
layout(yaxis = list(title = "Price"))
# plot volume bar chart
pp <- df %>%
plot_ly(x=~Date, y=~AAPL.Volume, type='bar', name = "AAPL Volume",
color = ~direction, colors = c('#17BECF','#7F7F7F')) %>%
layout(yaxis = list(title = "Volume"))
# create rangeselector buttons
rs <- list(visible = TRUE, x = 0.5, y = -0.055,
xanchor = 'center', yref = 'paper',
font = list(size = 9),
buttons = list(
list(count=1,
label='RESET',
step='all'),
list(count=1,
label='1 YR',
step='year',
stepmode='backward'),
list(count=3,
label='3 MO',
step='month',
stepmode='backward'),
list(count=1,
label='1 MO',
step='month',
stepmode='backward')
))
# subplot with shared x axis
p <- subplot(p, pp, heights = c(0.7,0.2), nrows=2,
shareX = TRUE, titleY = TRUE) %>%
layout(title = paste("Apple: 2015-02-14 -",Sys.Date()),
xaxis = list(rangeselector = rs),
legend = list(orientation = 'h', x = 0.5, y = 1,
xanchor = 'center', yref = 'paper',
font = list(size = 10),
bgcolor = 'transparent'))
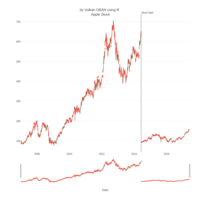
quantmod and plotly
library(plotly)
library(quantmod)
getSymbols("AAPL",src='yahoo')
df <- data.frame(Date=index(AAPL),coredata(AAPL))
# annotation
a <- list(text = "Stock Split",
x = '2014-06-06',
y = 1.02,
xref = 'x',
yref = 'paper',
xanchor = 'left',
showarrow = FALSE
)
# use shapes to create a line
l <- list(type = line,
x0 = '2014-06-06',
x1 = '2014-06-06',
y0 = 0,
y1 = 1,
xref = 'x',
yref = 'paper',
line = list(color = 'black',
width = 0.5)
)
p <- df %>%
plot_ly(x = ~Date, type="candlestick",
open = ~AAPL.Open, close = ~AAPL.Close,
high = ~AAPL.High, low = ~AAPL.Low) %>%
layout(title = "Apple Stock",
annotations = a,
shapes = l)
quantmod
getSymbols("AAPL")
chartSeries(AAPL)
title(" quantmod ", sub = "",
cex.main = 1, font.main= 2, col.main= "green",
cex.sub = 0.75, font.sub =1, col.sub = "red")

GGally
a<- ggpairs(iris)
a

psych package
pairs.panels(iris[1:4],bg=c("red","purple","blue")[iris$Species],pch=21,main=" Fisher Iris data by Species",hist.col="purple")
igraph
Show in New WindowClear OutputExpand/Collapse Output
shiny.tag
shiny.tag
shiny.tag
shiny.tag
shiny.tag
shiny.tag
shiny.tag
shiny.tag
Show in New WindowClear OutputExpand/Collapse Output
Error: unexpected symbol in:
"
print(p)Show"
Modify Chunk OptionsRun All Chunks AboveRun Current ChunkModify Chunk OptionsRun All Chunks AboveRun Current ChunkModify Chunk OptionsRun All Chunks AboveRun Current Chunk
Console~/
> library(miniCRAN)
> library(igraph)
>
>
> pk <- c("igraph","agop","bc3net","BDgraph","c3net","camel",
+ "cccd", "CDVine", "CePa", "CINOEDV", "cooptrees","corclass", "cvxclustr", "dcGOR",
+ "ddepn","dils", "dnet", "dpa", "ebdbNet", "editrules",
+ "fanovaGraph", "fastclime", "FisHiCal",
+ "flare", "G1DBN", "gdistance", "GeneNet", "GeneReg", "genlasso", "ggm", "gRapfa", "hglasso",
+ "huge", "igraphtosonia", "InteractiveIGraph", "iRefR", "JGL", "lcd", "linkcomm", "locits",
+ "loe", "micropan", "mlDNA", "mRMRe", "nets", "netweavers", "optrees", "packdep", "PAGI",
+ "pathClass", "PBC", "phyloTop", "picasso", "PoMoS", "popgraph", "PROFANCY", "qtlnet", "RCA",
+ "ReliabilityTheory", "rEMM", "restlos", "rgexf", "RNetLogo", "ror", "RWBP", "sand", "SEMID",
+ "shp2graph", "SINGLE", "spacejam", "TDA", "timeordered", "tnet")
>
>
> dg <- makeDepGraph(pk)
> plot(dg,main=" Network of reverse depends for igraph",cex=.4,vertex.size=8)

Plot
> require(graphics)
>
> fr <- function(x) { ## Rosenbrock Banana function
+ x1 <- x[1]
+ x2 <- x[2]
+ 100 * (x2 - x1 * x1)^2 + (1 - x1)^2
+ }
> grr <- function(x) { ## Gradient of 'fr'
+ x1 <- x[1]
+ x2 <- x[2]
+ c(-400 * x1 * (x2 - x1 * x1) - 2 * (1 - x1),
+ 200 * (x2 - x1 * x1))
+ }
> optim(c(-1.2,1), fr)
> (res <- optim(c(-1.2,1), fr, grr, method = "BFGS"))
> optimHess(res$par, fr, grr)
> optim(c(-1.2,1), fr, NULL, method = "BFGS", hessian = TRUE)
> ## These do not converge in the default number of steps
> optim(c(-1.2,1), fr, grr, method = "CG")
> optim(c(-1.2,1), fr, grr, method = "CG", control = list(type = 2))
> optim(c(-1.2,1), fr, grr, method = "L-BFGS-B")
>
> flb <- function(x)
+ { p <- length(x); sum(c(1, rep(4, p-1)) * (x - c(1, x[-p])^2)^2) }
> ## 25-dimensional box constrained
> optim(rep(3, 25), flb, NULL, method = "L-BFGS-B",
+ lower = rep(2, 25), upper = rep(4, 25)) # par[24] is *not* at boundary
>
> ## "wild" function , global minimum at about -15.81515
> fw <- function (x)
+ 10*sin(0.3*x)*sin(1.3*x^2) + 0.00001*x^4 + 0.2*x+80
> plot(fw, -50, 50, n = 1000, main = "optim() minimising 'wild function'")
>
> res <- optim(50, fw, method = "SANN",
+ control = list(maxit = 20000, temp = 20, parscale = 20))
> res
> ## Now improve locally {typically only by a small bit}:
> (r2 <- optim(res$par, fw, method = "BFGS"))
> points(r2$par, r2$value, pch = 8, col = "red", cex = 2)
>
> ## Combinatorial optimization: Traveling salesman problem
> library(stats) # normally loaded
>
> eurodistmat <- as.matrix(eurodist)
>
> distance <- function(sq) { # Target function
+ sq2 <- embed(sq, 2)
+ sum(eurodistmat[cbind(sq2[,2], sq2[,1])])
+ }
>
> genseq <- function(sq) { # Generate new candidate sequence
+ idx <- seq(2, NROW(eurodistmat)-1)
+ changepoints <- sample(idx, size = 2, replace = FALSE)
+ tmp <- sq[changepoints[1]]
+ sq[changepoints[1]] <- sq[changepoints[2]]
+ sq[changepoints[2]] <- tmp
+ sq
+ }
>
> sq <- c(1:nrow(eurodistmat), 1) # Initial sequence: alphabetic
> distance(sq)
[1] 29625
> # rotate for conventional orientation
> loc <- -cmdscale(eurodist, add = TRUE)$points
> x <- loc[,1]; y <- loc[,2]
> s <- seq_len(nrow(eurodistmat))
> tspinit <- loc[sq,]
>
> plot(x, y, type = "n", asp = 1, xlab = "", ylab = "",
+ main = "prepared by Volkan OBAN using R stats package
+ optim \n initial solution of traveling salesman problem", axes = FALSE)
> arrows(tspinit[s,1], tspinit[s,2], tspinit[s+1,1], tspinit[s+1,2],
+ angle = 10, col = "green")
> text(x, y, labels(eurodist), cex = 0.8)
>
> set.seed(123) # chosen to get a good soln relatively quickly
> res <- optim(sq, distance, genseq, method = "SANN",
+ control = list(maxit = 30000, temp = 2000, trace = TRUE,
+ REPORT = 500))
> tspres <- loc[res$par,]
> plot(x, y, type = "n", asp = 1, xlab = "", ylab = "",
+ main = "prepared by Volkan OBAN using R stats package optim \n optim() 'solving' traveling salesman problem", axes = FALSE)
> arrows(tspres[s,1], tspres[s,2], tspres[s+1,1], tspres[s+1,2],
+ angle = 10, col = "red")
> text(x, y, labels(eurodist), cex = 0.8)
>

ggfortify
http://www.sthda.com/english/wiki/ggfortify-extension-to-ggplot2-to-handle-some-popular-packages-r-software-and-data-visualization

ggfortify
http://www.sthda.com/english/wiki/ggfortify-extension-to-ggplot2-to-handle-some-popular-packages-r-software-and-data-visualization

sunshine
> par(mar=c(0,0,0,0))
> pie(abs(rnorm(150)) , radius=10 , border="transparent" , xlim=c(0,5) )

latticeExtra package
> xyplot(stl(log(co2), s.window=21),
+ main = "STL decomposition of CO2 data")

ggplot2 - waffle chart
library(ggplot2)
# Here's some data I had lying around
tb <- structure(list(region = c("Africa", "Asia", "Latin America",
"Other", "US-born"), ncases = c(36L, 34L, 56L, 2L, 44L)), .Names = c("region",
"ncases"), row.names = c(NA, -5L), class = "data.frame")
# A bar chart of counts
ggplot(tb, aes(x = region, weight = ncases, fill = region)) +
geom_bar()
# Bar chart of percentages
ggplot(tb, aes(x = region, weight = ncases/sum(ncases), fill = region)) +
geom_bar() +
scale_y_continuous(formatter = 'percent')
# Pie chart equivalents. Forgive me, Hadley, for I must sin.
ggplot(tb, aes(x = factor(1), weight = ncases, fill = region)) +
geom_bar(width = 1) +
coord_polar(theta = "y") +
labs(x = "", y = "")
ggplot(tb, aes(x = factor(1), weight = ncases/sum(ncases), fill = region)) +
geom_bar() +
scale_y_continuous(formatter = 'percent') +
coord_polar(theta = "y") +
labs(x = "", y = "")
# Waffles
# How many rows do you want the y axis?
ndeep <- 5
# I need to convert my data into a data.frame with a unique specified x
# and y axis for each case
# Note - it's actually important to specify y first for a
# horizontally-accumulating waffle
tb4waffles <- expand.grid(y = 1:ndeep,
x = seq_len(ceiling(sum(tb$ncases) / ndeep)))
# Expand the counts into a full vector of region labels - i.e., de-aggregate
regionvec <- rep(tb$region, tb$ncases)
# Depending on the value of ndeep, there might be more spots on the x-y grid
# than there are cases - so fill those with NA
tb4waffles$region <- c(regionvec, rep(NA, nrow(tb4waffles) - length(regionvec)))
# Plot it
ggplot(tb4waffles, aes(x = x, y = y, fill = region)) +
geom_tile(color = "white") + # The color of the lines between tiles
scale_fill_manual("Region of Birth",
values = RColorBrewer::brewer.pal(5, "Dark2")) +
opts(title = "TB Cases by Region of Birth")

waffle chart-waffle package
http://harrycaufield.net/severalog/2016/7/29/8jt1lrt7hd2vqh4heu7mcuqrdcg0od

waffle chart
ref. and code: http://harrycaufield.net/severalog/2016/7/29/8jt1lrt7hd2vqh4heu7mcuqrdcg0od
network package
> data(flo)
> nflo<-network(flo)
> #Display the network, indicating degree and flagging the Medicis
> plot(nflo, vertex.cex=apply(flo,2,sum)+1, usearrows=FALSE,vertex.sides=3+apply(flo,2,sum),vertex.col=2+(network.vertex.names(nflo)=="Medici"))
plotly network viz.
> library(plotly)
> library(igraph)
>
> data(karate, package="igraphdata")
> G <- upgrade_graph(karate)
> L <- layout.circle(G)
> vs <- V(G)
> es <- as.data.frame(get.edgelist(G))
>
> Nv <- length(vs)
> Ne <- length(es[1]$V1)
> Xn <- L[,1]
> Yn <- L[,2]
>
> network <- plot_ly(x = ~Xn, y = ~Yn, mode = "markers", text = vs$label, hoverinfo = "text")
> edge_shapes <- list()
> for(i in 1:Ne) {
+ v0 <- es[i,]$V1
+ v1 <- es[i,]$V2
+
+ edge_shape = list(
+ type = "line",
+ line = list(color = "#030303", width = 0.3),
+ x0 = Xn[v0],
+ y0 = Yn[v0],
+ x1 = Xn[v1],
+ y1 = Yn[v1]
+ )
+
+ edge_shapes[[i]] <- edge_shape
+ }
> axis <- list(title = "", showgrid = FALSE, showticklabels = FALSE, zeroline = FALSE)
>
> p <- layout(
+ network,
+ title = 'by Volkan OBAN using R - igraph \n Karate Network',
+ shapes = edge_shapes,
+ xaxis = axis,
+ yaxis = axis
+ )
> p

likert
require(likert)
> data(pisaitems)
>
> ##### Item 24: Reading Attitudes
> items24 <- pisaitems[,substr(names(pisaitems), 1,5) == 'ST24Q']
>
> items24 <- rename(items24, c(
+ ST24Q01="I read only if I have to.",
+ ST24Q02="Reading is one of my favorite hobbies.",
+ ST24Q03="I like talking about books with other people.",
+ ST24Q04="I find it hard to finish books.",
+ ST24Q05="I feel happy if I receive a book as a present.",
+ ST24Q06="For me, reading is a waste of time.",
+ ST24Q07="I enjoy going to a bookstore or a library.",
+ ST24Q08="I read only to get information that I need.",
+ ST24Q09="I cannot sit still and read for more than a few minutes.",
+ ST24Q10="I like to express my opinions about books I have read.",
+ ST24Q11="I like to exchange books with my friends."))
> l24g <- likert(items24[,1:2], grouping=pisaitems$CNT)
> plot(l24g)
heart.
> dat<- data.frame(t=seq(0, 2*pi, by=0.1) )
> xhrt <- function(t) 16*sin(t)^3
> yhrt <- function(t) 13*cos(t)-5*cos(2*t)-2*cos(3*t)-cos(4*t)
> dat$y=yhrt(dat$t)
> dat$x=xhrt(dat$t)
> with(dat, plot(x,y, type="l"))
> with(dat, polygon(x,y, col="darkred"))
BAMMtools package
ixx <- rep(c(10, 30, 40), 2);
plot.new()
par(mfrow=c(2,3));
colschemes <- list();
colschemes[1:3] <- 'temperature'
colschemes[4:6] <- list(c('blue', 'gray', 'red'))
for (i in 1:length(ixx)) {
par(mar=c(0,0,0,0))
index <- ixx[i]
eventsub <- subsetEventData(edata_whales, index=index);
plot.bammdata(eventsub, method='polar', pal= colschemes[[i]], par.reset=FALSE, lwd=3)
addBAMMshifts(eventsub, method='polar', index=1, col='white', bg='black', cex=5, par.reset=FALSE)
}
BAMMtools package
library(BAMMtools)
data(whales, events.whales)
edata_whales <- getEventData(whales, events.whales, burnin=0.1)
plot.bammdata(edata_whales, lwd=3, method="polar", pal="temperature")
data(primates, events.primates)
ed <- getEventData(primates, events.primates, burnin=0.25, type = 'trait')
par(mfrow=c(1,3), mar=c(1, 0.5, 0.5, 0.5), xpd=TRUE)
q <- plot.bammdata(ed, tau=0.001, breaksmethod='linear', lwd=2)
addBAMMshifts(ed, par.reset=FALSE, cex=2)
title(sub='linear',cex.sub=2, line=-1)
addBAMMlegend(q, location=c(0, 1, 140, 220))
q <- plot.bammdata(ed, tau=0.001, breaksmethod='linear', color.interval=c(NA,0.12), lwd=2)
addBAMMshifts(ed, par.reset=FALSE, cex=2)
title(sub='linear - color.interval',cex.sub=2, line=-1)
addBAMMlegend(q, location=c(0, 1, 140, 220))
q <- plot.bammdata(ed, tau=0.001, breaksmethod='jenks', lwd=2)
addBAMMshifts(ed, par.reset=FALSE, cex=2)
title(sub='jenks',cex.sub=2, line=-1)
addBAMMlegend(q, location=c(0, 1, 140, 220))

geomnet ggnetwork
> library(ggnetwork)
> set.seed(10312016)
> ggplot(ggnetwork(em.net, arrow.gap = 0.02, layout = "fruchtermanreingold"),
+ aes(x, y, xend = xend, yend = yend)) +
+ geom_edges(
+ aes(color = curr_empl_type),
+ alpha = 0.25,
+ arrow = arrow(length = unit(5, "pt"),
+ type = "closed"),
+ curvature = 0.05) +
+ geom_nodes(aes(color = curr_empl_type),
+ size = 4) +
+ scale_color_brewer("Employment Type",
+ palette = "Set1") +
+ theme_blank() +
+ theme(legend.position = "bottom")

Plot
library(tidyverse)
library(rvest)
library(magrittr)
library(ggmap)
library(stringr)
ref:https://www.r-bloggers.com/how-to-make-a-global-map-in-r-step-by-step/

heatmap.2
library(gplots)
>
> #Build the matrix data to look like a correlation matrix
> x <- matrix(rnorm(64), nrow=8)
> x <- (x - min(x))/(max(x) - min(x)) #Scale the data to be between 0 and 1
> for (i in 1:8) x[i, i] <- 1.0 #Make the diagonal all 1's
>
> #Format the data for the plot
> xval <- formatC(x, format="f", digits=2)
> pal <- colorRampPalette(c(rgb(0.96,0.96,1), rgb(0.1,0.1,0.9)), space = "rgb")
>
> #Plot the matrix
> x_hm <- heatmap.2(x, Rowv=FALSE, Colv=FALSE, dendrogram="none", main="by Volkan OBAN using R \n 8 X 8 Matrix Using Heatmap.2", xlab="Columns", ylab="Rows", col=pal, tracecol="#303030", trace="none", cellnote=xval, notecol="black", notecex=0.8, keysize = 1.3, margins=c(5, 5))

netdiffudeR package
set.seed(1231)
# Random scale-free diffusion network
x <- rdiffnet(1000, 4, seed.graph="scale-free", seed.p.adopt = .025,
rewire = FALSE, seed.nodes = "central",
rgraph.arg=list(self=FALSE, m=4),
threshold.dist = function(id) runif(1,.2,.4))
# Diffusion map (no random toa)
dm0 <- diffusionMap(x, kde2d.args=list(n=150, h=1), layout=igraph::layout_with_fr)
# Random
diffnet.toa(x) <- sample(x$toa, size = nnodes(x))
# Diffusion map (random toa)
dm1 <- diffusionMap(x, layout = dm0$coords, kde2d.args=list(n=150, h=.5))
oldpar <- par(no.readonly = TRUE)
col <- colorRampPalette(blues9)(100)
par(mfrow=c(1,2), oma=c(1,0,0,0), cex=.8)
image(dm0, col=col, main="Non-random Times of Adoption\nAdoption from the core.")
image(dm1, col=col, main="Random Times of Adoption")
par(mfrow=c(1,1))
mtext("Both networks have the same distribution on times of adoption", 1,
outer = TRUE)
sna package in R
g<-matrix(0,50,50)
g[1,]<-1; g[,1]<-1 #Create a star
gplot(g)
gplot(rewire.ws(g,0.05))
sna package in R
gplot(rgws(1,100,1,2,1))

arulesViz
library(arules)
> rules.all <- apriori(titanic.raw)
> load("titanic.raw.rdata")
> library(arulesViz)
> plot(rules.all)
plot(rules.all,main=" ", method = "graph", control = list(type = "items"))
networks
> net.bg <- sample_pa(80)
>
> V(net.bg)$size <- 8
>
> V(net.bg)$frame.color <- "firebrick3"
>
> V(net.bg)$color <- "hotpink"
>
> V(net.bg)$label <- ""
> l <- layout_in_circle(net.bg)
>
> plot(net.bg)
geomnet -- ggmap
metro_map <- ggmap::get_map(location = c(left = -77.22257, bottom = 39.05721,
right = -77.11271, top = 39.14247))
ggmap::ggmap(metro_map) +
geom_net(data = tripnet, layout.alg = NULL, labelon = TRUE,
vjust = -0.5, ealpha = 0.5,
aes(from_id = from_id,
to_id = to_id,
x = long, y = lat,
linewidth = n / 15,
colour = Metro)) +
scale_colour_manual("Metro Station", values = c("grey40", "darkorange")) +
theme_net() %+replace% theme(aspect.ratio=NULL, legend.position = "bottom") +
coord_map()
ref:https://cran.r-project.org/web/packages/ggCompNet/vignettes/examples-from-paper.html

Plot
> data(bikes, package = 'geomnet')
> # data step for geomnet
> tripnet <- fortify(as.edgedf(bikes$trips), bikes$stations[,c(2,1,3:5)])
> tripnet$Metro = FALSE
> idx <- grep("Metro", tripnet$from_id)
> tripnet$Metro[idx] <- TRUE
>
> # plot the bike sharing network shown in Figure 7b
> set.seed(1232016)
> ggplot(aes(from_id = from_id, to_id = to_id), data = tripnet) +
+ geom_net(aes(linewidth = n / 15, colour = Metro),
+ labelon = TRUE, repel = TRUE) +
+ theme_net() +
+ xlim(c(-0.1, 1.1)) +
+ scale_colour_manual("Metro Station", values = c("grey40", "darkorange")) +
+ theme(legend.position = "bottom")

geomnet and ggplot2
data(football, package = 'geomnet')
rownames(football$vertices) <-
football$vertices$label
# create network
fb.net <- network::network(football$edges[, 1:2],
directed = TRUE)
# create node attribute (what conference is team in?)
fb.net %v% "conf" <-
football$vertices[
network.vertex.names(fb.net), "value"
]
# create edge attribute (between teams in same conference?)
network::set.edge.attribute(
fb.net, "same.conf",
football$edges$same.conf)
set.seed(5232011)
ggnet2(fb.net, mode = "fruchtermanreingold",
color = "conf", palette = "Paired",
color.legend = "Conference",
edge.color = c("color", "grey75"))
---
ftnet <- fortify(as.edgedf(football$edges), football$vertices)
ftnet$schools <- ifelse(
ftnet$value == "Independents", ftnet$from_id, "")
# create data plot
set.seed(5232011)
ggplot(data = ftnet,
aes(from_id = from_id, to_id = to_id)) +
geom_net(layout.alg = 'fruchtermanreingold',
aes(colour = value, group = value,
linetype = factor(same.conf != 1),
label = schools),
linewidth = 0.5,
size = 5, vjust = -0.75, alpha = 0.3) +
theme_net() +
theme(legend.position = "bottom") +
scale_colour_brewer("Conference", palette = "Paired") +
guides(linetype = FALSE)

ggnet and ggplot2
> library(ggnet)
> data(email, package = 'geomnet')
>
> # create node attribute data
> em.cet <- as.character(
+ email$nodes$CurrentEmploymentType)
> names(em.cet) = email$nodes$label
>
> # remove the emails sent to all employees
> edges <- subset(email$edges, nrecipients < 54)
> # create network
> em.net <- edges[, c("From", "to") ]
> em.net <- network::network(em.net, directed = TRUE)
> # create employee type node attribute
> em.net %v% "curr_empl_type" <-
+ em.cet[ network.vertex.names(em.net) ]
> set.seed(10312016)
> ggnet2(em.net, color = "curr_empl_type",
+ size = 4, palette = "Set1",
+ arrow.size = 5, arrow.gap = 0.02,
+ edge.alpha = 0.25, mode = "fruchtermanreingold",
+ edge.color = c("color", "grey50"),
+ color.legend = "Employment Type") + ggtitle("by Volkan OBAN using R - ggnet") +
+ theme(legend.position = "bottom")
> email$edges <- email$edges[, c(1,5,2:4,6:9)]
> emailnet <- fortify(
+ as.edgedf(subset(email$edges, nrecipients < 54)),
+ email$nodes)
> set.seed(10312016)
> ggplot(data = emailnet,
+ aes(from_id = from_id, to_id = to_id)) +
+ geom_net(layout.alg = "fruchtermanreingold",
+ aes(colour = CurrentEmploymentType,
+ group = CurrentEmploymentType,
+ linewidth = 3 * (...samegroup.. / 8 + .125)),
+ ealpha = 0.25,
+ size = 4, curvature = 0.05,
+ directed = TRUE, arrowsize = 0.5) +
+ scale_colour_brewer("Employment Type", palette = "Set1") +
+ theme_net() + ggtitle("by Volkan OBAN using R - ggnet") +
+ theme(legend.position = "bottom")
> set.seed(10312016)
> ggplot(data = emailnet,
+ aes(from_id = from_id, to_id = to_id)) +
+ geom_net(layout.alg = "fruchtermanreingold",
+ aes(colour = CurrentEmploymentType,
+ group = CurrentEmploymentType,
+ linewidth = 3 * (...samegroup.. / 8 + .125)),
+ ealpha = 0.25,
+ size = 4, curvature = 0.05,
+ directed = TRUE, arrowsize = 0.5) +
+ scale_colour_brewer("Employment Type", palette = "Set1") +
+ theme_net() +
+ theme(legend.position = "bottom")
>

geomnet
> library(geomnet)
> data(madmen, package = "geomnet")
>
> # code for geom_net
> # data step: merge edges and nodes by the "from" column
>
> MMnet <- fortify(as.edgedf(madmen$edges), madmen$vertices)
set.seed(10052016)
ggplot(data = MMnet, aes(from_id = from_id, to_id = to_id)) +
geom_net(aes(colour = Gender), layout.alg = "kamadakawai",
size = 2, labelon = TRUE, vjust = -0.6, ecolour = "grey60",
directed =FALSE, fontsize = 3, ealpha = 0.5) +
scale_colour_manual(values = c("#FF69B4", "#0099ff")) +
xlim(c(-0.05, 1.05)) +
theme_net() +
theme(legend.position = "bottom")
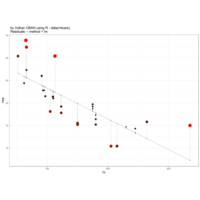
Residuals
fit <- lm(mpg ~ hp, data = mtcars)
d <- mtcars
fit <- lm(mpg ~ hp, data = d)
d$predicted <- predict(fit) # Save the predicted values
d$residuals <- residuals(fit) # Save the residual values
# Quick look at the actual, predicted, and residual values
library(dplyr)
d %>% select(mpg, predicted, residuals) %>% head()
ggplot(d, aes(x = hp, y = mpg)) +
geom_smooth(method = "lm", se = FALSE, color = "lightgrey") +
geom_segment(aes(xend = hp, yend = predicted), alpha = .2) +
# > Color adjustments made here...
geom_point(aes(color = abs(residuals))) + # Color mapped to abs(residuals)
scale_color_continuous(low = "black", high = "red") + # Colors to use here
guides(color = FALSE) + # Color legend removed
# <
geom_point(aes(y = predicted), shape = 1) +
theme_bw()
and
// another visualization
ggplot(d, aes(x = hp, y = mpg)) +
geom_smooth(method = "lm", se = FALSE, color = "lightgrey") +
geom_segment(aes(xend = hp, yend = predicted), alpha = .2) +
# > Color AND size adjustments made here...
geom_point(aes(color = abs(residuals), size = abs(residuals))) + # size also mapped
scale_color_continuous(low = "black", high = "red") +
guides(color = FALSE, size = FALSE) + # Size legend also removed
# <
geom_point(aes(y = predicted), shape = 1) +
theme_bw()
chorddiagram
library(dplyr)
titanic_tbl <- dplyr::tbl_df(Titanic)
titanic_tbl <- titanic_tbl %>%
mutate_each(funs(factor), Class:Survived)
by_class_survival <- titanic_tbl %>%
group_by(Class, Survived) %>%
summarize(Count = sum(n))
titanic.mat <- matrix(by_class_survival$Count, nrow = 4, ncol = 2)
dimnames(titanic.mat ) <- list(Class = levels(titanic_tbl$Class),
Survival = levels(titanic_tbl$Survived))
print(titanic.mat)
groupColors <- c("#2171b5", "#6baed6", "#bdd7e7", "#bababa", "#d7191c", "#1a9641")
chorddiag(titanic.mat, type = "bipartite",
groupColors = groupColors,
tickInterval = 50)
circos
library(migest)
demo(cfplot_nat, package = "migest", ask = FALSE)
circos
library("migest")
demo(cfplot_reg2, package = "migest", ask = FALSE)
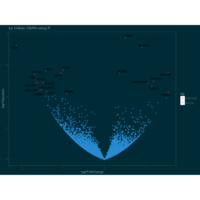
Plot
library(dplyr)
> library(ggplot2)
>
> # Read data from the web
> url = "https://gist.githubusercontent.com/stephenturner/806e31fce55a8b7175af/raw/1a507c4c3f9f1baaa3a69187223ff3d3050628d4/results.txt"
>
> results = read.table(url, header=TRUE)
> results = mutate(results, sig=ifelse(results$padj<0.05, "FDR<0.05", "Not Sig"))
>
> p = ggplot(results, aes(log2FoldChange, -log10(pvalue))) +
+ geom_point(aes(col=sig)) + ggtitle("by Volkan OBAN using R") +
+ scale_color_manual(values=c("darkblue", "purple"))
> p
> p+geom_text(data=filter(results, padj<0.05), aes(label=Gene))
> library(ggrepel)
>
> p+geom_text_repel(data=filter(results, padj<0.05), aes(label=Gene))
> library(ggthemes)
> library(ggrepel)
>
> p+geom_text_repel(data=filter(results, padj<0.05), aes(label=Gene)) + theme_wsj() + scale_colour_wsj("colors6", "")
or
> p+geom_text_repel(data=filter(results, padj<0.05), aes(label=Gene)) + theme_solarized(light = FALSE) +
+ scale_colour_solarized("red")
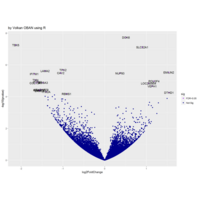
ggplot2
library(dplyr)
library(ggplot2)
# Read data from the web
url = "https://gist.githubusercontent.com/stephenturner/806e31fce55a8b7175af/raw/1a507c4c3f9f1baaa3a69187223ff3d3050628d4/results.txt"
results = read.table(url, header=TRUE)
results = mutate(results, sig=ifelse(results$padj<0.05, "FDR<0.05", "Not Sig"))
p = ggplot(results, aes(log2FoldChange, -log10(pvalue))) +
geom_point(aes(col=sig)) +
scale_color_manual(values=c("red", "black"))
p
p+geom_text(data=filter(results, padj<0.05), aes(label=Gene))

Boxplot for Time Series
code:
library(RColorBrewer)
# Create Data
days=rep(c("monday","tuesday","wenesday","thursday","friday","saturday","sunday") , each=120)
time=rep (rep( paste(seq(0,22,2),seq(2,24,2),sep="-") , each=10 ) , 7)
value=rep ( rep(seq(0,22,2) , each=10 ) , 7)+rnorm(mean=10, sd=10 , length(time))
data=data.frame(days, time, value)
# Create a color palette
my_colors = brewer.pal(9, "Blues")
my_colors = colorRampPalette(my_colors)(12)
# Make the boxplot
boxplot(data$value ~ data$time+data$days , xaxt="n" , xlab="" , col=my_colors , pch=20 , cex=0.3 , ylab="value" )
abline(v= seq(0, 12*7, 12) +0.5 , col="grey")
axis(1, labels=unique(days) , at=seq(6,12*7,12) )
# Add general trend
a=aggregate(data$value , by=list(data$time, data$days) , mean)
lines(a[,3], type="l" , col="red" , lwd=2)

rcharts
ref. and codes: http://timelyportfolio.blogspot.com.tr/2013/06/r-plotting-financial-time-series.html
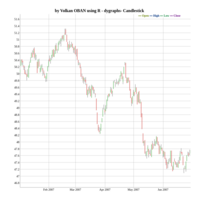
dygraphs
> library(dygraphs)
> dygraph(ldeaths) %>%
+ dyRangeSelector() %>%
+ dyUnzoom()
> library(xts)
> data(sample_matrix)
> library(dygraphs)
> dygraph(sample_matrix) %>%
+ dyCandlestick()
> library(xts)
> data(sample_matrix)
> library(dygraphs)
> dygraph(sample_matrix, main = "by Volkan OBAN using R - dygraphs- Candlestick") %>%
dyCandlestick()

dygraphs
library(quantmod)
library(dygraphs)
tickers <- c("AAPL", "MSFT")
getSymbols(tickers)
closePrices <- do.call(merge, lapply(tickers, function(x) Cl(get(x))))
dateWindow <- c("2008-01-01", "2009-01-01")
dygraph(closePrices, main = "Value", group = "stock") %>%
dyRebase(value = 100) %>%
dyRangeSelector(dateWindow = dateWindow)
dygraph(closePrices, main = "Percent", group = "stock") %>%
dyRebase(percent = TRUE) %>%
dyRangeSelector(dateWindow = dateWindow)
dygraph(closePrices, main = "None", group = "stock") %>%
dyRangeSelector(dateWindow = dateWindow)
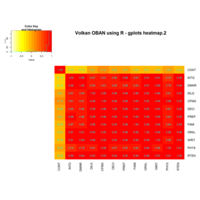
heatmap.2
> data(USJudgeRatings)
> symnum( cU <- cor(USJudgeRatings) )
hM <- format(round(cU, 2))
> hM
heatmap.2(cU, Rowv=FALSE,main=" Volkan OBAN using R - gplots heatmap.2", symm=TRUE, col=rev(heat.colors(16)),
+ distfun=function(c) as.dist(1 - c), trace="none",
+ cellnote=hM)

harmonograph
f1=jitter(sample(c(2,3),1));f2=jitter(sample(c(2,3),1));f3=jitter(sample(c(2,3),1));f4=jitter(sample(c(2,3),1))
d1=runif(1,0,1e-02);d2=runif(1,0,1e-02);d3=runif(1,0,1e-02);d4=runif(1,0,1e-02)
p1=runif(1,0,pi);p2=runif(1,0,pi);p3=runif(1,0,pi);p4=runif(1,0,pi)
xt = function(t) exp(-d1*t)*sin(t*f1+p1)+exp(-d2*t)*sin(t*f2+p2)
yt = function(t) exp(-d3*t)*sin(t*f3+p3)+exp(-d4*t)*sin(t*f4+p4)
t=seq(1, 100, by=.001)
dat=data.frame(t=t, x=xt(t), y=yt(t))
with(dat, plot(x,y, type="l", xlim =c(-2,2), ylim =c(-2,2), xlab = "", ylab = "", xaxt='n', yaxt='n'))
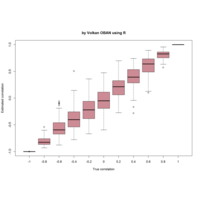
BoxPlot
> library(mvtnorm)
> k <- 100 # Number of samples for each correlation
> N <- 20 # Size of the samples
> r <- seq(-1, 1, by=.2) # The true correlations
> n <- length(r)
> rr <- matrix(NA, nr=n, nc=k)
> for (i in 1:n) {
+ for (j in 1:k) {
+ x <- rmvnorm(N, sigma=matrix(c(1, r[i], r[i], 1), nr=2, nc=2))
+ rr[i,j] <- cor( x[,1], x[,2] )
+ }
+ }
> estimated.correlation <- as.vector(rr)
> true.correlation <- r[row(rr)]
> boxplot(estimated.correlation ~ true.correlation,
+ col = "purple",
+ xlab = "True correlation", main="y Volkan OBAN using R",
+ ylab = "Estimated correlation" )
> library(mvtnorm)
> k <- 100 # Number of samples for each correlation
> N <- 20 # Size of the samples
> r <- seq(-1, 1, by=.2) # The true correlations
> n <- length(r)
> rr <- matrix(NA, nr=n, nc=k)
> for (i in 1:n) {
+ for (j in 1:k) {
+ x <- rmvnorm(N, sigma=matrix(c(1, r[i], r[i], 1), nr=2, nc=2))
+ rr[i,j] <- cor( x[,1], x[,2] )
+ }
+ }
> estimated.correlation <- as.vector(rr)
> true.correlation <- r[row(rr)]
> boxplot(estimated.correlation ~ true.correlation,
+ col = "lightpink3",
+ xlab = "True correlation", main="by Volkan OBAN using R",
+ ylab = "Estimated correlation" )

geom_boxplot() + facet_wrap(~ ) ggplot2
ggplot(diamonds, aes(x = cut, y = price, fill = clarity)) +
+ geom_boxplot() +
+ facet_wrap(~ clarity, scale = "free")

geom_boxplot() + facet_wrap(~ ) ggplot2
> library(ggplot2)
>
> # create fake dataset with additional attributes - sex, sample, and temperature
> x <- data.frame(values = c(runif(100, min = 0), runif(100), runif(100, max = 3), runif(100)),
letter = rep(c('o', 'v'), each = 100),
sample = rep(c('VVV', 'OOO'), each = 200),
s = sample(c('1984', '1990', '2000', '2019'), 400, replace = TRUE) )
>
>
> ggplot(x, aes(x = sample, y = values, fill = letter)) +
+ geom_boxplot() +
+ facet_wrap(~ s)

ggplot2 facet_wrap
> p<- ggplot(diamonds, aes(x=cut, y=price, fill=cut))
> p + geom_boxplot() + facet_wrap(~clarity, scales="free")

ggplot2
require (ggplot2)
> require (plyr)
> library(reshape2)
>
> set.seed(1234)
> x<- rnorm(100)
> y.1<-rnorm(80)
> y.2<-rnorm(60)
> y.3<-rnorm(75)
> y.4<-rnorm(105)
> y.5<-rnorm(80)
> y.6<-rnorm(90)
> df<- (as.data.frame(cbind(x,y.1,y.2,y.3,y.4,y.5,y.6)))
ggplot(dfmelt, aes(value, x, group = round_any(x, 0.5), fill=variable))+
+ geom_boxplot() +
+ geom_jitter() +
+ facet_wrap(~variable)
threejs
N <- 100
i <- sample(3, N, replace=TRUE)
x <- matrix(rnorm(N*3),ncol=3)
lab <- c("small", "bigger", "biggest")
scatterplot3js(x, color=rainbow(N), labels=lab[i], size=i, renderer="canvas")

threejs
> data(flights)
> # Approximate locations as factors
> dest <- factor(sprintf("%.2f:%.2f",flights[,3], flights[,4]))
> # A table of destination frequencies
> freq <- sort(table(dest), decreasing=TRUE)
> # The most frequent destinations in these data, possibly hub airports?
> frequent_destinations <- names(freq)[1:10]
> # Subset the flight data by destination frequency
> idx <- dest %in% frequent_destinations
> frequent_flights <- flights[idx, ]
> # Lat/long and counts of frequent flights
> ll <- unique(frequent_flights[,3:4])
> # Plot frequent destinations as bars, and the flights to and from
> # them as arcs. Adjust arc width and color by frequency.
> globejs(lat=ll[,1], long=ll[,2], arcs=frequent_flights,
+ arcsHeight=0.3, arcsLwd=2, arcsColor="#FFFFFF", arcsOpacity=0.15,
+ atmosphere=TRUE, color="#000080", pointsize=0.5)
>
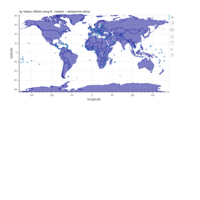
rbokeh
> library(maps)
> data(world.cities)
> caps <- subset(world.cities, capital == 1)
> caps$population <- prettyNum(caps$pop, big.mark = ",")
> figure(width = 800, height = 450,title="by Volkan OBAN using R - rbokeh -- data(world.cities)", padding_factor = 0) %>%
+ ly_map("world", col = "darkblue") %>%
+ ly_points(long, lat, data = caps, size = 5,
+ hover = c(name, country.etc, population))

wordcloud
library(wordcloud)
>
> #Create a list of words (Random words concerning my work)
> a=c("VOLKAN OBAN","Mathematics","Data Science","Machine Learning","scikit-learn","solution","MLib","Apache Spark","Analysis","Big Data","Science","Statistics","Data", "Programming","ggplot2","matplotlib-seaborn","Volkan","VOLKAN","Istanbul","kNN","R", "R","Data-Viz","Python","kmeans","Programming","Graph Theory ","Operations Research", "Predictive Analytics","Clustering","Data Science","Prescriptive Analytics","Analytics","Classification")
>
> #I give a frequency to each word of this list
> b=sample(seq(0,1,0.01) , length(a) , replace=TRUE)
> par(bg="deeppink4")
> wordcloud(a , b , col=terrain.colors(length(a) , alpha=0.9) , rot.per=0.3 )
art in R. ref: Gaston Sanchez
x = seq(-50, 50, by = 1)
y = -(x^2)
# set graphic parameters
op = par(bg = 'black', mar = rep(0.5, 4))
# Plot
plot(y, x, type = 'n')
lines(y, x, lwd = 2*runif(1), col = hsv(0.08, 1, 1, alpha = runif(1, 0.5, 0.9)))
for (i in seq(10, 2500, 10))
{
lines(y-i, x, lwd = 2*runif(1), col = hsv(0.08, 1, 1, alpha = runif(1, 0.5, 0.9)))
}
for (i in seq(500, 600, 10))
{
lines(y - i, x, lwd = 2*runif(1), col = hsv(0, 1, 1, alpha = runif(1, 0.5, 0.9)))
}
for (i in seq(2000, 2300, 10))
{
lines(y - i, x, lwd = 2*runif(1), col = hsv(0, 1, 1, alpha = runif(1, 0 .5, 0.9)))
}
for (i in seq(100, 150, 10))
{
lines(y - i, x, lwd = 2*runif(1), col = hsv(0, 1, 1, alpha = runif(1, 0.5, 0.9)))
}
# signature
legend("bottomright", legend="© Gaston Sanchez", bty = "n", text.col="gray70")
Plot
library(RColorBrewer)
>
> # Classic palette BuPu, with 4 colors
> coul = brewer.pal(4, "BuPu")
>
> # I can add more tones to this palette :
> coul = colorRampPalette(coul)(25)
>
> # Plot it
> pie(rep(1, length(coul)), col = coul , main=" R - piechart - RColorBrewer ")

Plot3D package
require(plot3D)
Zorunlu paket yükleniyor: plot3D
> lon <- seq(165.5, 188.5, length.out = 30)
> lat <- seq(-38.5, -10, length.out = 30)
> xy <- table(cut(quakes$long, lon),
+ cut(quakes$lat, lat))
> xmid <- 0.5*(lon[-1] + lon[-length(lon)])
> ymid <- 0.5*(lat[-1] + lat[-length(lat)])
>
> par (mar = par("mar") + c(0, 0, 0, 2))
> hist3D(x = xmid, y = ymid, z = xy,
+ zlim = c(-20, 40), main = " Earth quakes",
+ ylab = "latitude", xlab = "longitude",
+ zlab = "counts", bty= "g", phi = 5, theta = 25,
+ shade = 0.2, col = "white", border = "black",
+ d = 1, ticktype = "detailed")
>
> with (quakes, scatter3D(x = long, y = lat,
+ z = rep(-20, length.out = length(long)),
+ colvar = quakes$depth, col = gg.col(100),
+ add = TRUE, pch = 18, clab = c("depth", "m"),
+ colkey = list(length = 0.5, width = 0.5,
+ dist = 0.05, cex.axis = 0.8, cex.clab = 0.8)
+ ))
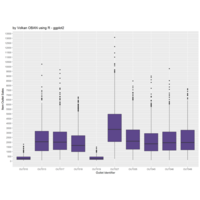
ggplot2
ggplot(train, aes(Outlet_Identifier, Item_Outlet_Sales)) + geom_boxplot(fill = "mediumpurple4")+
+ scale_y_continuous("Item Outlet Sales", breaks= seq(0,15000, by=500))+
+ labs(title = " R - ggplot2", x = "Outlet Identifier")
data:https://docs.google.com/spreadsheets/d/1PR5StHxg2jlMCb4IUilGSEwhylXn-3q3EJucSaVolCU/edit#gid=0

scatterplot
train<-read.csv(mart.csv)
Error in read.table(file = file, header = header, sep = sep, quote = quote, :
object 'mart.csv' not found
> train <- read.csv(file="mart.csv", header=TRUE, sep=",")
> ggplot(train, aes(Item_Visibility, Item_MRP)) + geom_point(aes(color = Item_Type)) +
+ scale_x_continuous("Item Visibility", breaks = seq(0,0.35,0.05))+
+ scale_y_continuous("Item MRP", breaks = seq(0,270,by = 30))+
+ theme_bw()
data:https://docs.google.com/spreadsheets/d/1PR5StHxg2jlMCb4IUilGSEwhylXn-3q3EJucSaVolCU/edit#gid=0

ggplot2
ref: https://www.r-bloggers.com/improved-net-stacked-distribution-graphs-via-ggplot2-trickery/
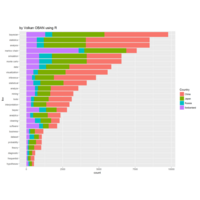
ggplot2
library("ggplot2")
> data <- read.csv("ggplot-data.csv", header=TRUE, nrows=200)
> gg <- ggplot(data, aes(x=Keyword))
> gg <- gg + geom_bar(aes(weight=Traffic, fill=Country) + coord_flip()
+ )
> gg
> data$kw <- reorder(data$Keyword, data$Traffic)
> gg <- ggplot(data, aes(x=kw))
>
> gg <- gg + geom_bar(aes(weight=Traffic, fill=Country)) + coord_flip()
>
> gg
> gg <- ggplot(data, aes(x=kw))
>
> gg <- gg + geom_bar(aes(weight=Traffic, fill=Country)) + coord_flip()
>
> gg

ggplot2 facet_wrap
> c <- ggplot(diamonds, aes(clarity, fill=cut)) + geom_bar()
> c + facet_wrap(~cut, scales = "free_y") + coord_flip(
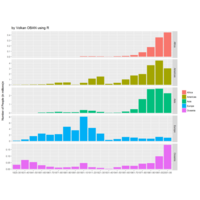
ggplot2
library(ggplot2)
> df <- structure(c(106487, 495681, 1597442,
+ 2452577, 2065141, 2271925, 4735484, 3555352,
+ 8056040, 4321887, 2463194, 347566, 621147,
+ 1325727, 1123492, 800368, 761550, 1359737,
+ 1073726, 36, 53, 141, 41538, 64759, 124160,
+ 69942, 74862, 323543, 247236, 112059, 16595,
+ 37028, 153249, 427642, 1588178, 2738157,
+ 2795672, 2265696, 11951, 33424, 62469,
+ 74720, 166607, 404044, 426967, 38972, 361888,
+ 1143671, 1516716, 160037, 354804, 996944,
+ 1716374, 1982735, 3615225, 4486806, 3037122,
+ 17, 54, 55, 210, 312, 358, 857, 350, 7368,
+ 8443, 6286, 1750, 7367, 14092, 28954, 80779,
+ 176893, 354939, 446792, 33333, 69911, 53144,
+ 29169, 18005, 11704, 13363, 18028, 46547,
+ 14574, 8954, 2483, 14693, 25467, 25215,
+ 41254, 46237, 98263, 185986), .Dim = c(19,
+ 5), .Dimnames = list(c("1820-30", "1831-40",
+ "1841-50", "1851-60", "1861-70", "1871-80",
+ "1881-90", "1891-00", "1901-10", "1911-20",
+ "1921-30", "1931-40", "1941-50", "1951-60",
+ "1961-70", "1971-80", "1981-90", "1991-00",
+ "2001-06"), c("Europe", "Asia", "Americas",
+ "Africa", "Oceania")))
> library(reshape)
Attaching package: ‘reshape’
The following objects are masked from ‘package:plyr’:
rename, round_any
The following object is masked from ‘package:Matrix’:
expand
> df.m <- melt(df)
> df.m <- rename(df.m, c(X1 = "Period", X2 = "Region"))
a <- ggplot(df.m, aes(x = Period, y = value/1e+06,
+ fill = Region)) + options(title = "Migration to the United States by Source Region (1820-2006)") +
+ labs(x = NULL, y = "Number of People (in millions)n",
+ fill = NULL)
> b <- a + geom_bar(stat = "identity", position = "stack")
> b
c <- b+ facet_grid(Region ~ .) + options(legend.position = "none")
> c
> total <- cast(df.m, Period ~ ., sum)
> total <- rename(total, c(`(all)` = "value"))
> total$Region <- "Total"
> df.m.t <- rbind(total, df.m)
> c1 <- c %+% df.m
> total <- cast(df.m, Period ~ ., sum)
> total <- rename(total, c(`(all)` = "value"))
> total$Region <- "Total"
> df.m.t <- rbind(total, df.m)
> c1 <- c %+% df.m
> c1
> c2 <- c1 + facet_grid(Region ~ ., scale = "free_y")
> c2
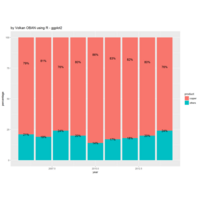
ggplot2
ibrary(ggplot2)
> library(ggthemes)
> library(extrafont)
Registering fonts with R
> library(plyr)
Attaching package: ‘plyr’
The following object is masked from ‘package:network’:
is.discrete
> library(scales)
Attaching package: ‘scales’
The following object is masked _by_ ‘.GlobalEnv’:
cscale
> charts.data <- read.csv("data.csv")
> p <- ggplot() + geom_bar(aes(y = percentage, x = year, fill = product), data = charts.data,stat="identity")
p <- p + geom_text(data=charts.data, aes(x = year, y = percentage,
+ label = paste0(percentage,"%")), size=4)
p
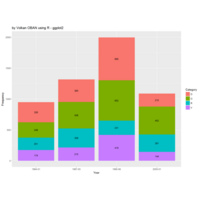
ggplot2
Year <- c(rep(c("1984-01", "1987-05", "1990-06", "2005-01"), each = 4))
Category <- c(rep(c("V", "O", "R", "D"), times = 4))
Frequency <- c(174, 248, 201, 326, 215, 428, 309, 365, 419, 652, 231, 695, 144, 452, 281, 210)
Data <- data.frame(Year, Category, Frequency)
ggplot(Data, aes(x = Year, y = Frequency, fill = Category, label = Frequency)) +
geom_bar(stat = "identity") +
geom_text(size = 3, position = position_stack(vjust = 0.5))

ggplot2 facet_grid
> ggplot(diamonds, aes(clarity)) +
+ geom_bar(aes(fill = cut)) +
+ facet_grid(cut ~ .)
Gauge Chart in R
ref and code :https://www.r-bloggers.com/gauge-chart-in-r/
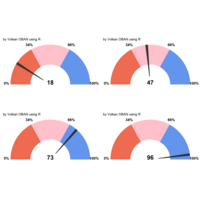
gauge
gg.gauge <- function(pos,breaks=c(0,42,58,100)) {
+ require(ggplot2)
+ get.poly <- function(a,b,r1=0.5,r2=1.0) {
+ th.start <- pi*(1-a/100)
+ th.end <- pi*(1-b/100)
+ th <- seq(th.start,th.end,length=100)
+ x <- c(r1*cos(th),rev(r2*cos(th)))
+ y <- c(r1*sin(th),rev(r2*sin(th)))
+ return(data.frame(x,y))
+ }
+ ggplot()+ ggtitle("by Volkan OBAN using R \n Gauge") +
+ geom_polygon(data=get.poly(breaks[1],breaks[2]),aes(x,y),fill="green")+
+ geom_polygon(data=get.poly(breaks[2],breaks[3]),aes(x,y),fill="pink")+
+ geom_polygon(data=get.poly(breaks[3],breaks[4]),aes(x,y),fill="purple")+
+ geom_polygon(data=get.poly(pos-1,pos+1,0.2),aes(x,y))+
+ geom_text(data=as.data.frame(breaks), size=5, fontface="bold", vjust=0,
+ aes(x=1.1*cos(pi*(1-breaks/100)),y=1.1*sin(pi*(1-breaks/100)),label=paste0(breaks,"%")))+
+ annotate("text",x=0,y=0,label=pos,vjust=0,size=8,fontface="bold")+
+ coord_fixed()+
+ theme_bw()+
+ theme(axis.text=element_blank(),
+ axis.title=element_blank(),
+ axis.ticks=element_blank(),
+ panel.grid=element_blank(),
+ panel.border=element_blank())
+ }
> gg.gauge(52,breaks=c(0,42,58,100)
+
+ )
> library(gridExtra)
> grid.newpage()
> grid.draw(arrangeGrob(gg.gauge(22),gg.gauge(36),
+ gg.gauge(71),gg.gauge(95),ncol=2))
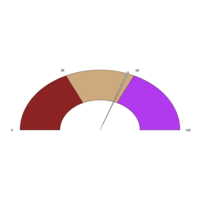
gauge
gg.gauge <- function(pos,breaks=c(0,42,58,100)) {
+ require(ggplot2)
+ get.poly <- function(a,b,r1=0.5,r2=1.0) {
+ th.start <- pi*(1-a/100)
+ th.end <- pi*(1-b/100)
+ th <- seq(th.start,th.end,length=100)
+ x <- c(r1*cos(th),rev(r2*cos(th)))
+ y <- c(r1*sin(th),rev(r2*sin(th)))
+ return(data.frame(x,y))
+ }
+ ggplot()+ ggtitle("by Volkan OBAN using R \n Gauge") +
+ geom_polygon(data=get.poly(breaks[1],breaks[2]),aes(x,y),fill="green")+
+ geom_polygon(data=get.poly(breaks[2],breaks[3]),aes(x,y),fill="pink")+
+ geom_polygon(data=get.poly(breaks[3],breaks[4]),aes(x,y),fill="purple")+
+ geom_polygon(data=get.poly(pos-1,pos+1,0.2),aes(x,y))+
+ geom_text(data=as.data.frame(breaks), size=5, fontface="bold", vjust=0,
+ aes(x=1.1*cos(pi*(1-breaks/100)),y=1.1*sin(pi*(1-breaks/100)),label=paste0(breaks,"%")))+
+ annotate("text",x=0,y=0,label=pos,vjust=0,size=8,fontface="bold")+
+ coord_fixed()+
+ theme_bw()+
+ theme(axis.text=element_blank(),
+ axis.title=element_blank(),
+ axis.ticks=element_blank(),
+ panel.grid=element_blank(),
+ panel.border=element_blank())
+ }
> gg.gauge(52,breaks=c(0,42,58,100)
+
+ )

DiagrammeR
> spec <- "
+ digraph { 'VOLKAN OBAN \n Data Scientist ' }
+ [1]: LETTERS[1]
+ "
>
>
> grViz(replace_in_spec(spec))
DiagrammeR
> spec <- "
+ digraph { '@1' }
+ [1]: LETTERS[1]
+ "
> grViz(replace_in_spec(spec))
> spec <- "
+ digraph a_nice_graph {
+ node [fontname = Arial]
+ a [label = 'by VOLKAN OBAN using R ']
+ b [label = 'Mathematics']
+ c [label = 'Data Science']
+ d [label = 'Analytics']
+ e [label = 'Programming']
+ f [label = 'Machine Learning']
+ g [label = 'Python']
+ h [label = 'Statistics']
+ i [label = 'R']
+ j [label = 'Istanbul']
+ a -> { b c d e f g h i j}
+ }
+ [1]: 'top'
+ [2]: 10:20
+ "
> grViz(replace_in_spec(spec))
>

ggplot2 and ggthemr
> ggthemr('lilac')
>ggplot(data = mpg, mapping = aes(x = class, y = hwy)) +
+ geom_boxplot() +
+ coord_flip() +

ggplot2 and ggthemr
sea
ggplot(data = mpg, mapping = aes(x = class, y = hwy)) +
+ geom_boxplot() +
+ coord_flip()

ggplot2 and ggthemr
....................
ggplot(data = diamonds) +
geom_bar(mapping = aes(x = cut, fill = clarity), position = "dodge")

ggplot2 and ggthemr
> ggthemr('lilac')
> ggplot(data = diamonds) +
+ geom_bar(mapping = aes(x = cut, fill = clarity), position = "dodge") + ggtitle("by Volkan OBAN using R - ggplot2 and ggthemr packages /data(diamonds)")

ggplot2 and ggthemr
> ggthemr('sea')
> ggplot(data = diamonds) +
+ geom_bar(mapping = aes(x = cut, fill = clarity), position = "dodge") + ggtitle("by Volkan OBAN using R - ggplot2 and ggthemr packages /data(diamonds)")

Visualize kmeans clustering
ref: http://handsondatascience.com/ClustersO.pdf

ggmap
> ds<-map_data("world")
> p <- ggplot(ds, aes( x=long, y=lat, group=group))
> p <-p + geom_polygon() + ggtitle("by Volkan OBAN using R - ggmap")
> p
>
> p <- ggplot(ds, aes(x=long, y=lat, group=group, fill=region))
> p <- p + geom_polygon()
> p <- p + geom_polygon()
> p <- p + theme(legend.position = "none")
> p

Visualize kmeans clustering
> library(rattle) # Load weather dataset. Normalise names normVarNames().
Rattle: A free graphical interface for data mining with R.
Version 4.1.0 Copyright (c) 2006-2015 Togaware Pty Ltd.
Type 'rattle()' to shake, rattle, and roll your data.
> library(randomForest) # Impute missing using na.roughfix().
randomForest 4.6-12
Type rfNews() to see new features/changes/bug fixes.
> # Identify the dataset.
> dsname <- "weather"
> ds <- get(dsname)
> names(ds) <- normVarNames(names(ds))
> vars <- names(ds)
> target <- "rain_tomorrow"
> risk <- "risk_mm"
> id <- c("date", "location")
> # Ignore the IDs and the risk variable.
> ignore <- union(id, if (exists("risk")) risk)
> # Ignore variables which are completely missing.
> mvc <- sapply(ds[vars], function(x) sum(is.na(x))) # Missing value count.
> mvn <- names(ds)[(which(mvc == nrow(ds)))] # Missing var names.
> ignore <- union(ignore, mvn)
> # Initialise the variables
> vars <- setdiff(vars, ignore)
> # Variable roles.
> inputc <- setdiff(vars, target)
> inputi <- sapply(inputc, function(x) which(x == names(ds)), USE.NAMES=FALSE)
> numi <- intersect(inputi, which(sapply(ds, is.numeric)))
> numc <- names(ds)[numi]
> cati <- intersect(inputi, which(sapply(ds, is.factor)))
> catc <- names(ds)[cati]
> # Impute missing values, but do this wisely - understand why missing.
> if (sum(is.na(ds[vars]))) ds[vars] <- na.roughfix(ds[vars])
> # Number of observations.
> nobs <- nrow(ds)
> model <- m.km <- kmeans(ds, 10)
> model <- m.kms <- kmeans(scale(ds[numi]), 10)
> model$size
[1] 34 54 15 70 24 32 30 44 43 20
> library(ggplot2)
> library(reshape)
Attaching package: ‘reshape’
The following object is masked from ‘package:Matrix’:
expand
> nclust <- 4
> model <- m.kms <- kmeans(scale(ds[numi]), nclust)
> dscm <- melt(model$centers)
> names(dscm) <- c("Cluster", "Variable", "Value")
> dscm$Cluster <- factor(dscm$Cluster)
> dscm$Order <- as.vector(sapply(1:length(numi), rep, nclust))
> p <- ggplot(dscm,
+ aes(x=reorder(Variable, Order),
+ y=Value, group=Cluster, colour=Cluster))
> p <- p + coord_polar()
> p <- p + geom_point()
> p <- p + geom_path()
> p <- p + labs(x=NULL, y=NULL)
> p <- p + theme(axis.ticks.y=element_blank(), axis.text.y = element_blank())
> p
>

Visualize kmeans clustering
> set.seed(32297)
d <- data.frame(x=runif(100),y=runif(100))
> clus <- kmeans(d,centers=5)
> d$cluster <- clus$cluster
> library('ggplot2')
> library('grDevices')
> h <- do.call(rbind,
+ lapply(unique(clus$cluster),
+ function(c) { f <- subset(d,cluster==c); f[chull(f),]}))
> ggplot() +
+ geom_text(data=d,aes(label=cluster,x=x,y=y,
+ color=cluster),size=3) +
+ geom_polygon(data=h,aes(x=x,y=y,group=cluster,fill=as.factor(cluster)),
+ alpha=0.4,linetype=0) +
+ theme(legend.position = "none")

wordcloud
> library(wordcloud)
>
> #Create a list of words (Random words concerning my work)
> a=c("Volkan OBAN","Clustering","Turkey","Istanbul","Classification","Istanbul Technical University","Mathematics",
+ "Data Science","Analysis","Machine Learning","Science","Statistics","Data",
+ "Programming","Clustering","Recommedation","Visualization","Spark","Business","VOLKAN","R", "R",
+ "Data-Viz","Python","Linux","Programming","Graphs","Numbers", "Big Data",
+ "Computing","Data-Science","Analytics","GitHub","OBAN")
>
> #I give a frequency to each word of this list
> b=sample(seq(0,1,0.01) , length(a) , replace=TRUE)
>
> #The package will automatically make the wordcloud ! (I add a black background)
> par(bg="hotpink4")
> wordcloud(a , b , col=terrain.colors(length(a) , alpha=0.9) , rot.per=0.3 )
>
Plot
> moxbuller = function(n) {
+ u = runif(n)
+ v = runif(n)
+ x = cos(2*pi*u)*sqrt(-2*log(v))
+ y = sin(2*pi*v)*sqrt(-2*log(u))
+ r = list(x=x, y=y)
+ return(r)
+ }
> r = moxbuller(50000)
> par(bg="aliceblue")
> par(mar=c(0,0,0,0))
> plot(r$x,r$y, pch=".", col="hotpink4",main=" \n by Volkan OBAN using R", cex=1.2)

Plot
library(magrittr)
>
> add_line_points2 <- function(plot, df, ...) {
+ plot +
+ geom_line(aes(x = Time, y = weight, group = Chick), ..., data = df) +
+ geom_point(aes(x = Time, y = weight), ..., data = df)
+ }
>
> (plot4 <- ggplot() %>% add_line_points2(diet1)
+ %>% add_line_points2(diet2, colour = "red")

Plot
> library(ggplot2)
>
> data(ChickWeight)
> diet1 <- subset(ChickWeight, Diet == 1)
> diet2 <- subset(ChickWeight, Diet == 2)
> add_line <- function(df) {
+ geom_line(aes(x = Time, y = weight, group = Chick), data = df)
+ }
>
> add_points <- function(df) {
+ geom_point(aes(x = Time, y = weight), data = df)
+ }
>
> add_line_points <- function(df) {
+ add_line(df) + add_points(df)
+ }
(p <- ggplot(aes(x = Time, y = weight, group = Chick, colour = Diet),
+ data = ChickWeight) +
+ geom_line() + geom_point())

Plot
library(ggplot2)
>
> data(ChickWeight)
> diet1 <- subset(ChickWeight, Diet == 1)
> diet2 <- subset(ChickWeight, Diet == 2)
> add_line <- function(df) {
+ geom_line(aes(x = Time, y = weight, group = Chick), data = df)
+ }
>
> add_points <- function(df) {
+ geom_point(aes(x = Time, y = weight), data = df)
+ }
>
> add_line_points <- function(df) {
+ add_line(df) + add_points(df)
p <- ggplot(aes(x = Time, y = weight, group = Chick), data = diet1) +
+ geom_line() + geom_point()
lattice package --wireframe and cloud
cloud(Sepal.Length ~ Petal.Length * Petal.Width | Species, data = iris,
screen = list(x = -90, y = 70),main="by Volkan OBAN using R", distance = .4, zoom = .6)

timeseries zoo package.
library(quantmod)
> tckrs <- c("SPY", "QQQ", "GDX", "DBO", "VWO")
> getSymbols(tckrs, from = "2007-01-01"
SPY.Close <- SPY[,4]
> QQQ.Close <- QQQ[,4]
> GDX.Close <- GDX[,4]
> DBO.Close <- DBO[,4]
> VWO.Close <- VWO[,4]
> SPY1 <- as.numeric(SPY.Close[1])
> QQQ1 <- as.numeric(QQQ.Close[1])
> GDX1 <- as.numeric(GDX.Close[1])
> DBO1 <- as.numeric(DBO.Close[1])
> VWO1 <- as.numeric(VWO.Close[1]
+ )
> SPY <- SPY.Close/SPY1
> QQQ <- QQQ.Close/QQQ1
> GDX <- GDX.Close/GDX1
> DBO <- DBO.Close/DBO1
> VWO <- VWO.Close/VWO1
> basket <- cbind(SPY, QQQ, GDX, DBO, VWO
+ )
> zoo.basket <- as.zoo(basket
+ )
> tsRainbow <- rainbow(ncol(zoo.basket))
> # Plot the overlayed series
> plot(x = zoo.basket, ylab = "Cumulative Return", main = "by Volkan OBAN using R \n Cumulative Returns",
+ col = tsRainbow, screens = 1)
> # Set a legend in the upper left hand corner to match color to return series
> legend(x = "topleft", legend = c("SPY", "QQQ", "GDX", "DBO", "VWO"),
lty = 1,col = tsRainbow)
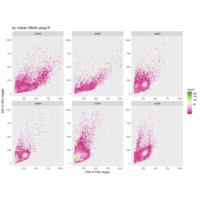
ggcyto from bioconductor
> library(ggcyto)
> data(GvHD)
> fs <- GvHD[subset(pData(GvHD), Patient %in%5:7 & Visit %in% c(5:6))[["name"]]]
> fr <- fs[[1]]
> p <- ggcyto(fs, aes(x = `FSC-H`))
> p <- ggcyto(fs, aes(x = `FSC-H`, y = `SSC-H`)) + ggtitle("by Volkan OBAN using R")
> p <- p + geom_hex(bins = 128)
> p
Warning message:
Removed 257 rows containing missing values (geom_hex).
> p + scale_fill_gradientn(colours = rainbow(7), trans = "sqrt")
> library(knitr)
> library(RColorBrewer)
> p + scale_fill_gradientn(colours = brewer.pal(n=8,name="PiYG"),trans="sqrt")
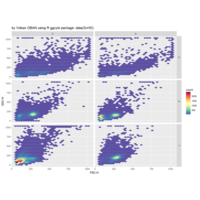
ggcyto from bioconductor
library(ggcyto)
data(GvHD)
fs <- GvHD[subset(pData(GvHD), Patient %in%5:7 & Visit %in% c(5:6))[["name"]]]
fr <- fs[[1]]
p1 <- ggplot(mapping = aes(x = `FSC-H`, y = `SSC-H`)) + myColor_scale_fill + facet_grid(Patient~Visit)
p1 + stat_binhex(data = fs, bin = 64)

ggplot2 and ggthemes
> p<-ggplot(diamonds, aes(cut, price)) +
+ geom_boxplot() +
+ coord_flip() + theme_solarized() +
+ scale_colour_solarized("purple") + ggtitle("by Volkan OBAN using R \n data(diamonds) ") + theme(plot.title = element_text(size = 12, face = "bold")
+ )
> p

SVM plot
> data(iris)
> m2 <- svm(Species~., data = iris)
> plot(m2, iris, Petal.Width ~ Petal.Length,
+ slice = list(Sepal.Width = 3, Sepal.Length = 4))
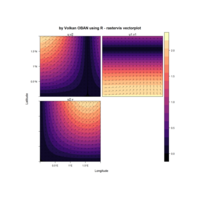
rasterVis
u1 <- cos(y) * cos(x)
v1 <- cos(y) * sin(x)
u2 <- sin(y) * sin(x)
v2 <- sin(y) * cos(x)
field <- stack(u, u1, u2, v, v1, v2)
names(field) <- c('u', 'u1', 'u2', 'v', 'v1', 'v2')
vectorplot(field, isField='dXY',
narrows=300, lwd.arrows=.4,
par.settings=BTCTheme(),
layout=c(3, 1))
## uLayer and vLayer define which layers contain
## horizontal and vertical components, respectively
vectorplot(field, isField='dXY',
narrows=300,
uLayer=1:3,
vLayer=6:4)

rasterVis
u1 <- cos(y) * cos(x)
v1 <- cos(y) * sin(x)
u2 <- sin(y) * sin(x)
v2 <- sin(y) * cos(x)
field <- stack(u, u1, u2, v, v1, v2)
names(field) <- c('u', 'u1', 'u2', 'v', 'v1', 'v2')
vectorplot(field, isField='dXY',
narrows=300, lwd.arrows=.4,
par.settings=BTCTheme(),
layout=c(3, 1))
## uLayer and vLayer define which layers contain
## horizontal and vertical components, respectively
vectorplot(field, isField='dXY',
narrows=300,
uLayer=1:3,
vLayer=6:4)
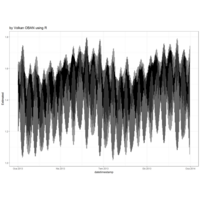
SWMPr and oce
library(SWMPr)
library(oce)
# clean input data, one hour time step, subset, fill gaps
dat <- qaqc(apadbwq) %>%
setstep(timestep = 60) %>%
subset(., subset = c('2013-01-01 0:0', '2013-12-31 0:0'), select = 'depth') %>%
na.approx(maxgap = 1e6)
# get model
datsl <- as.sealevel(elevation = dat$depth, time = dat$datetimestamp)
mod <- tidem(t = datsl)
# add predictions to observed data
dat$Estimated <- predict(mod)
# plot
ggplot(dat, aes(x = datetimestamp, y = Estimated)) +
geom_line() +
theme_bw()

Plot
constituents <- c('M2', 'S2', 'N2', 'K2', 'K1', 'O1', 'P1')
# loop through tidal components, predict each with tidem
preds <- sapply(constituents, function(x){
mod <- tidem(t = datsl, constituent = x)
pred <- predict(mod)
pred - mean(pred)
})
# combine prediction, sum, add time data
predall <- rowSums(preds) + mean(datsl[['elevation']])
preds <- data.frame(time = datsl[['time']], preds, Estimated = predall)
head(preds)
mod <- tidem(t = datsl)
Note: the record is too short to fit for constituents: SA PI1 S1 PSI1 GAM2 H1 H2 T2 R2
>
> # get components of interest
> amps <- data.frame(mod@data[c('name', 'amplitude')]) %>%
+ filter(name %in% constituents) %>%
+ arrange(amplitude)
> amps
name amplitude
1 K2 0.01091190
2 N2 0.01342395
3 S2 0.02904518
4 P1 0.04100388
5 O1 0.11142455
6 M2 0.12005114
7 K1 0.12865764
> dat$Estimated <- predict(mod)
>
> # plot one month
> ggplot(dat, aes(x = datetimestamp, y = depth)) +
+ geom_point() +
+ geom_line(aes(y = Estimated), colour = 'blue') +
+ scale_x_datetime(limits = as.POSIXct(c('2013-07-01', '2013-07-31'))) +
+ scale_y_continuous(limits = c(0.9, 2)) +
+ theme_bw()

SWMPr and oce
library(SWMPr)
Warning message:
package ‘SWMPr’ was built under R version 3.3.3
> library(oce)
>
> # clean, one hour time step, subset, fill gaps
> dat <- qaqc(apadbwq) %>%
+ setstep(timestep = 60) %>%
+ subset(subset = c('2013-01-01 0:0', '2013-12-31 0:0'), select = 'depth') %>%
+ na.approx(maxgap = 1e6)
> datsl <- as.sealevel(elevation = dat$depth, time = dat$datetimestamp)
> plot(datsl,main="by Volkan OBAN using R")
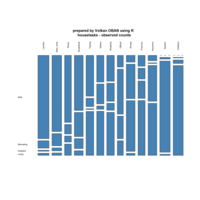
mosaic plot
> library("graphics")
> # Mosaic plot of observed values
> mosaicplot(housetasks, las=2, col="steelblue",
+ main = " \n housetasks - observed counts")

MAPS
> require(maps)
> Tur = map_data('world', region = 'Turkey')
> ggplot(Tur, aes(x = long, y = lat, group = group)) +
+ geom_polygon(fill = 'red', colour = 'black') +ggtitle("TURKEY- TÜRKİYE CENNETİM"

Plot
> c <- ggplot(diamonds, aes(carat, price))
> c + geom_bin2d()
> require(hexbin)
> c + geom_hex()
> c + geom_hex(bins = 10)

ggplot2
> wdata = data.frame(
+ s = factor(rep(c("F", "M"), each=200)),
+ weight = c(rnorm(200, 55), rnorm(200, 58)))
a <- ggplot(wdata, aes(x = weight))
> a + geom_dotplot()

ggplot2
> set.seed(1234)
> wdata = data.frame(
+ s = factor(rep(c("F", "M"), each=200)),
+ weight = c(rnorm(200, 55), rnorm(200, 58)))
> head(wdata)
s weight
1 F 53.79293
2 F 55.27743
3 F 56.08444
4 F 52.65430
5 F 55.42912
6 F 55.50606
> qplot(s, weight, data = wdata, geom = "dotplot",
+ stackdir = "center", binaxis = "y", dotsize = 0.5)
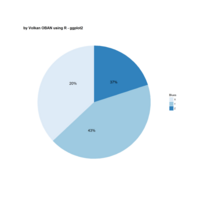
pie chart
> df <- data.frame(
+ group = c("X", "Y", "Z"),
+ value = c(37, 43, 20)
+ )
> head(df)
group value
1 X 37
2 Y 43
3 Z 20
> library(ggplot2)
> bp<- ggplot(df, aes(x="", y=value, fill=group))+
+ geom_bar(width = 1, stat = "identity")
> bp
> bp<- ggplot(df, aes(x="", y=value, fill=group))+
+ geom_bar(width = 1, stat = "identity")
> pie <- bp + coord_polar("y", start=0)
> pie
> pie + scale_fill_manual(values=c("#999999", "#E69F00", "#56B4E9"))
> ggplot(PlantGrowth, aes(x=factor(1), fill=group))+
+ geom_bar(width = 1)+
+ coord_polar("y")
> ggplot(PlantGrowth, aes(x=factor(1), fill=group))+
+ geom_bar(width = 1)+
+ coord_polar("y")
> blank_theme <- theme_minimal()+
+ theme(
+ axis.title.x = element_blank(),
+ axis.title.y = element_blank(),
+ panel.border = element_blank(),
+ panel.grid=element_blank(),
+ axis.ticks = element_blank(),
+ plot.title=element_text(size=14, face="bold")
+ )
> library(scales)
> pie + scale_fill_grey() + blank_theme +
+ theme(axis.text.x=element_blank()) +
+ geom_text(aes(y = value/3 + c(0, cumsum(value)[-length(value)]),
+ label = percent(value/100)), size=5
+ )
> pie + scale_fill_brewer("Blues") + blank_theme +
+ theme(axis.text.x=element_blank())+
+ geom_text(aes(y = value/3 + c(0, cumsum(value)[-length(value)]),
+ label = percent(value/100)), size=5)
>

ggplot2
correlation matrix
> mydata <- mtcars[, c(1,3,4,5,6,7)]
> cormat <- round(cor(mydata),2)
> library(reshape2)
> melted_cormat <- melt(cormat)
> head(melted_cormat)
library(ggplot2)
> ggplot(data = melted_cormat, aes(Var2, Var1, fill = value))+
geom_tile(color = "white")+
scale_fill_gradient2(low = "purple", high = "red", mid = "white",
midpoint = 0, limit = c(-1,1), space = "Lab",
name="Pearson\n Correlation") +
theme_minimal()+
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 12, hjust = 1))+
coord_fixed()

Plot
> ohio <- midwest %>%
+ filter(state == "OH") %>%
+ select(county, percollege) %>%
+ arrange(percollege) %>%
+ mutate(Avg = mean(percollege, na.rm = TRUE),
+ Above = ifelse(percollege - Avg > 0, TRUE, FALSE),
+ county = factor(county, levels = .$county)
ggplot(ohio, aes(percollege, county, color = Above)) +
+ geom_segment(aes(x = Avg, y = county, xend = percollege, yend = county), color = "grey50") + ggtitle("preprared by Volkan OBAN using R - ggplot2 - data(midwest) ") +
+ geom_point()

rworldmap
> library(rworldmap)
> newmap <- getMap(resolution = "high")
> plot(newmap,main=" R - rworldmap",
+ xlim = c(-20, 59),
+ ylim = c(35, 71),
+ asp = 1)

canvasXpress package
> data <- t(iris[,1:4])
> varAnnot <- as.matrix(iris[,5])
> colnames(varAnnot) <- "Species"
> canvasXpress(t(data),varAnnot=varAnnot, graphType='Scatter3D', colorBy='Species')

canvasXpress package
> data <- t(iris[,1:4])
> smpAnnot <- as.matrix(iris[,5])
> colnames(smpAnnot) <- "Species"
> canvasXpress(data,title="by Volkan OBAN using R \n canvasXpress package", smpAnnot=smpAnnot, graphType='Boxplot', groupingFactors=list('Species'))
> # or
> canvasXpress(data,title="by Volkan OBAN using R \n canvasXpress package",smpAnnot=smpAnnot, graphType='Boxplot', afterRender=list(list('groupSamples', list('Species'))))

ggplot2
> library(ggplot2)
> g <- ggplot(mpg, aes(manufacturer))
> g + geom_bar(aes(fill=class), width = 0.5) +
+ theme(axis.text.x = element_text(angle=65, vjust=0.6)) +
+ labs(title="by Volkan OBAN using R",
+ subtitle=" Categorywise Bar Chart \n Manufacturer of vehicles",
+ caption="Source: Manufacturers from 'mpg' dataset")
ggmap-İzmir
qmap(location = "izmir")
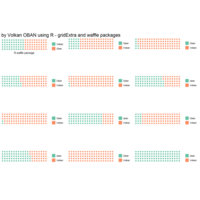
waffle and gridExtra
gridExtra::grid.arrange(
+ waffle(c(Volkan=50, Oban=50), rows=5,title="by Volkan OBAN using R - gridExtra and waffle packages", xlab="R-waffle package"),
+ waffle(c(Oban=25, Volkan=75), rows=5), waffle(c(Oban=7, Volkan=93), rows=5), waffle(c(Oban=42, Volkan=58), rows=5), waffle(c(Oban=2, Volkan=98), rows=5), waffle(c(oban=63, Volkan=37), rows=5),waffle(c(Oban=2, Volkan=98), rows=5), waffle(c(oban=75, Volkan=25), rows=5),waffle(c(Oban=15, Volkan=85), rows=5), waffle(c(oban=63, Volkan=37), rows=5),waffle(c(Oban=0, Volkan=100), rows=5), waffle(c(oban=100, Volkan=0), rows=5) )
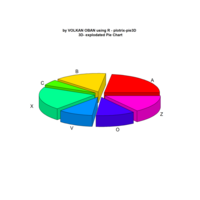
plotrix
> slices <- c(18, 12, 4, 16, 8, 9, 12)
> labels <- c("A", "B", "C", "X", "V", "O", "Z")
> library(plotrix)
> pie3D(slices,labels=labels,explode=0.1, main=" 3D- explodated Pie Chart")
ggraph
ref: https://www.r-bloggers.com/introduction-to-ggraph-layouts/

ggbeeswarm
> library(gridExtra)
> dat <- list( 'Normal'=rnorm(50),'Dense normal'= rnorm(500),'Bimodal'=c(rnorm(100), rnorm(100,5)), 'Trimodal'=c(rnorm(100), rnorm(100,5),rnorm(100,-3))
+ )
> labs<-rep(names(dat),sapply(dat,length))
> labs<-factor(labs,levels=unique(labs))
> dat<-unlist(dat)
>
>
>
>
>
> p1<-ggplot(mapping=aes(labs, dat)) +geom_quasirandom(method='smiley',alpha=.2) + ggtitle('Default (n/5)') + labs(x='Volkan OBAN')
> p2<-ggplot(mapping=aes(labs, dat)) + geom_quasirandom(method='smiley',nbins=50,alpha=.2) +ggtitle('nbins=50')
> p3<-ggplot(mapping=aes(labs, dat)) +geom_quasirandom(method='smiley',nbins=100,alpha=.2) + ggtitle('nbins=100')
> p4<-ggplot(mapping=aes(labs, dat)) +geom_quasirandom(method='smiley',nbins=250,alpha=.2) +ggtitle('nbins=250')
> grid.arrange(p1, p2, p3, p4, ncol=1)
>

psych package
ref:
https://cran.r-project.org/web/packages/psych/psych.pdf
factor Analysis- ggplot2 grid gridExtra and psych
ref: http://rpubs.com/danmirman/plotting_factor_analysis
ggtree
pp <- ggtree(tree) %>% phylopic("79ad5f09-cf21-4c89-8e7d-0c82a00ce728", print(pp)
ggtree
ref : https://bioconductor.org/packages/devel/bioc/manuals/ggtree/man/ggtree.pdf
mlrMBO
library(mlrMBO)
fun = makeSingleObjectiveFunction(
name = "SineMixture",
fn = function(x) sin(x[1])*cos(x[2])/2 + 0.04 * sum(x^2),
par.set = makeNumericParamSet(id = "x", len = 2, lower = -5, upper = 5)
)
ctrl = makeMBOControl()
# For this numeric optimization we are going to use the Expected Improvement as infill criterion:
ctrl = setMBOControlInfill(ctrl, crit = crit.ei)
# We will allow for exactly 25 evaluations of the objective function:
ctrl = setMBOControlTermination(ctrl, max.evals = 25L)
library(ggplot2)
des = generateDesign(n = 8L, par.set = getParamSet(fun), fun = lhs::randomLHS)
autoplot(fun, render.levels = TRUE) + geom_point(data = des)
mlrMBO ecr and plot3D
set.seed(1)
library(mlrMBO)
fun = makeSingleObjectiveFunction(
name = "SineMixture",
fn = function(x) sin(x[1])*cos(x[2])/2 + 0.04 * sum(x^2),
par.set = makeNumericParamSet(id = "x", len = 2, lower = -5, upper = 5)
)
library(plot3D)
plot3D(fun, contour = TRUE, lightning = TRUE)

ggforce
rocketData <- data.frame(
x = c(1,1,2,2),
y = c(1,2,2,3)
)
rocketData <- do.call(rbind, lapply(seq_len(500)-1, function(i) {
rocketData$y <- rocketData$y - c(0,i/500);
rocketData$group <- i+1;
rocketData
}))
rocketData2 <- data.frame(
x = c(2, 2.25, 2),
y = c(2, 2.5, 3)
)
rocketData2 <- do.call(rbind, lapply(seq_len(500)-1, function(i) {
rocketData2$x[2] <- rocketData2$x[2] - i*0.25/500;
rocketData2$group <- i+1 + 500;
rocketData2
}))
ggplot() + geom_link(aes(x=2, y=2, xend=3, yend=3, alpha=..index..,
size = ..index..), colour='goldenrod', n=500) +
geom_bezier(aes(x=x, y=y, group=group, colour=..index..),
data=rocketData) +
geom_bezier(aes(x=y, y=x, group=group, colour=..index..),
data=rocketData) +
geom_bezier(aes(x=x, y=y, group=group, colour=1),
data=rocketData2) +
geom_bezier(aes(x=y, y=x, group=group, colour=1),
data=rocketData2) +
geom_text(aes(x=1.65, y=1.65, label='vvv', angle=45),
colour='white', size=15) +
coord_fixed() +
scale_x_reverse() +
scale_y_reverse() +
scale_alpha(range=c(1, 0), guide='none') +
scale_size_continuous(range=c(20, 0.1), trans='exp',
guide='none') +
scale_color_continuous(guide='none') +
xlab('') + ylab('') +
ggtitle('ggforce: ggplot2') +
theme(plot.title = element_text(size = 20))

LDA-ggplot2
MASS package
data(iris)

corrplot
M <- cor(mtcars)
ord <- corrMatOrder(M, order = "AOE")
M2 <- M[ord,ord]
corrplot.mixed(M2)
corrplot.mixed(M2, lower = "ellipse", upper = "circle")
corrplot.mixed(M2, lower = "square", upper = "circle")
corrplot.mixed(M2, lower = "shade", upper = "circle")
corrplot.mixed(M2, tl.pos = "lt")
corrplot.mixed(M2, tl.pos = "lt", diag = "u")
corrplot.mixed(M2, tl.pos = "lt", , diag = "l")
corrplot
data(mtcars)
M <- cor(mtcars)
## different color series
col1 <- colorRampPalette(c("#7F0000","red","#FF7F00","yellow","white",
"cyan", "#007FFF", "blue","#00007F"))
col2 <- colorRampPalette(c("#67001F", "#B2182B", "#D6604D", "#F4A582", "#FDDBC7",
"#FFFFFF", "#D1E5F0", "#92C5DE", "#4393C3", "#2166AC", "#053061"))
col3 <- colorRampPalette(c("red", "white", "blue"))
col4 <- colorRampPalette(c("#7F0000","red","#FF7F00","yellow","#7FFF7F",
"cyan", "#007FFF", "blue","#00007F"))
wb <- c("white","black")
par(ask = TRUE)
## different color scale and methods to display corr-matrix
corrplot(M, method = "number", col = "black", cl.pos = "n")
corrplot(M, method = "number")
corrplot(M)
corrplot(M, order = "AOE")
corrplot(M, order = "AOE", addCoef.col = "grey")
corrplot(M, order = "AOE", col = col1(20), cl.length = 21, addCoef.col = "grey")
corrplot(M, order = "AOE", col = col1(10), addCoef.col = "grey")
corrplot(M, order = "AOE", col = col2(200))
corrplot(M, order = "AOE", col = col2(200), addCoef.col = "grey")
corrplot(M, order = "AOE", col = col2(20), cl.length = 21, addCoef.col = "grey")
corrplot(M, order = "AOE", col = col2(10), addCoef.col = "grey")

ggmap
> world <- map_data("world")
Attaching package: ‘maps’
The following object is masked from ‘package:plyr’:
ozone
> worldmap <- ggplot(world, aes(long, lat, group = group)) +
+ geom_path() +
+ scale_y_continuous(NULL, breaks = (-2:3) * 30, labels = NULL) +
+ scale_x_continuous(NULL, breaks = (-4:4) * 45, labels = NULL)
>
> worldmap + coord_map()
> # Some crazier projections
> worldmap + coord_map("ortho")
> worldmap + coord_map("stereographic")

ggmap
> world <- map_data("world")
Attaching package: ‘maps’
The following object is masked from ‘package:plyr’:
ozone
> worldmap <- ggplot(world, aes(long, lat, group = group)) +
+ geom_path() +
+ scale_y_continuous(NULL, breaks = (-2:3) * 30, labels = NULL) +
+ scale_x_continuous(NULL, breaks = (-4:4) * 45, labels = NULL)
>
> worldmap + coord_map()
> # Some crazier projections
> worldmap + coord_map("ortho")
> worldmap + coord_map("stereographic")

ggplot2
ref: http://jdobr.es/blog/data-vis-with-ggplot/

ggplot2
ref: http://jdobr.es/blog/data-vis-with-ggplot/
chemmineR package.
data(sdfsample)
(sdfset <- sdfsample)
## Plot single compound structure
plotStruc(sdfset[[1]])
## Plot several compounds structures
plot(sdfset[1:4])
## Highlighting substructures (here all rings)
myrings <- as.numeric(gsub(".*_", "", unique(unlist(rings(sdfset[1])))))
plot(sdfset[1], colbonds=myrings)
## Customize plot
plot(sdfset[1:4], griddim=c(2,2), print_cid=letters[1:4], print=FALSE, noHbonds=FALSE)
chemmineR package.
## Import SDFset sample set
data(sdfsample)
(sdfset <- sdfsample)
## Plot single compound structure
plotStruc(sdfset[[1]])
## Plot several compounds structures
plot(sdfset[1:4])
## Highlighting substructures (here all rings)
myrings <- as.numeric(gsub(".*_", "", unique(unlist(rings(sdfset[1])))))
plot(sdfset[1], colbonds=myrings)
chemmineR package.
data(sdfsample)
(sdfset <- sdfsample)
## Plot single compound structure
plotStruc(sdfset[[1]])
## Plot several compounds structures
plot(sdfset[1:4])
chemmineR package.
data(sdfsample)
sdfset <- sdfsample
## Create bond matrix for first two molecules in sdfset
conMA(sdfset[1:2], exclude=c("H"))
## Return bond matrix for first molecule and plot its structure with atom numbering
conMA(sdfset[[1]], exclude=c("H"))
plot(sdfset[1], atomnum = TRUE, noHbonds=FALSE , no_print_atoms = "", atomcex=0.8)
ref:https://www.bioconductor.org/packages/devel/bioc/manuals/ChemmineR/man/ChemmineR.pdf

grid package
dsmall <- diamonds[sample(nrow(diamonds), 1000), ]
> library(grid)
> a <- ggplot(dsmall, aes(color, price/carat)) + geom_jitter(size=4, alpha = I(1 / 1.5), aes(color=color))
> b <- ggplot(dsmall, aes(color, price/carat, color=color)) + geom_boxplot()
> c <- ggplot(dsmall, aes(color, price/carat, fill=color)) + geom_boxplot() + theme(legend.position = "none")
> grid.newpage() # Open a new page on grid device
> pushViewport(viewport(layout = grid.layout(2, 2))) # Assign to device viewport with 2 by 2 grid layout
> print(a, vp = viewport(layout.pos.row = 1, layout.pos.col = 1:2))
> print(b, vp = viewport(layout.pos.row = 2, layout.pos.col = 1))
> print(c, vp = viewport(layout.pos.row = 2, layout.pos.col = 2, width=0.3, height=0.3, x=0.8, y=0.8))
ggplot2
df <- data.frame(group = rep(c("Above", "Below"), each=10), x = rep(1:10, 2), y = c(runif(10, 0, 1), runif(10, -1, 0)))
> p <- ggplot(df, aes(x=x, y=y, fill=group)) + geom_bar(stat="identity", position="identity")
> p

ggplot2
ref: https://learnr.wordpress.com/page/4/
Dikesh Jariwala
Create Air Travel Route Maps in ggplot---R-bloggers
R-bloggers
# Read flight list
flights <- read.csv("flights.csv", stringsAsFactors = FALSE)
# Lookup coordinates
library(ggmap)
airports <- unique(c(flights$From, flights$To))
coords <- geocode(airports)
airports <- data.frame(airport=airports, coords)
flights <- merge(flights, airports, by.x="To", by.y="airport")
flights <- merge(flights, airports, by.x="From", by.y="airport")
# Plot flight routes
library(ggplot2)
library(ggrepel)
worldmap <- borders("world", colour="#efede1", fill="#efede1") # create a layer of borders
ggplot() + worldmap +
geom_curve(data=flights, aes(x = lon.x, y = lat.x, xend = lon.y, yend = lat.y), col = "#b29e7d", size = 1, curvature = .2) +
geom_point(data=airports, aes(x = lon, y = lat), col = "#970027") +
geom_text_repel(data=airports, aes(x = lon, y = lat, label = airport), col = "black", size = 2, segment.color = NA) +
theme(panel.background = element_rect(fill="white"),
axis.line = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_blank(),
axis.ticks = element_blank(),
axis.title.x = element_blank(),
axis.title.y = element_blank()
)

rAmcharts
Funnel
qgraph
qgraph(dat.3[-c(1:3,13:15),],layout=L.3,nNodes=30,directed=FALSE,edge.labels=TRUE,esize=14)
qgraph
> dat.3 <- matrix(c(1:15*2-1,1:15*2),,2)
> dat.3 <- cbind(dat.3,round(seq(-0.7,0.7,length=15),1))
> L.3 <- matrix(1:30,nrow=2)
> # Different esize:
> qgraph(dat.3,layout=L.3,directed=FALSE,edge.labels=TRUE,esize=14)
> qgraph(dat.3[-c(1:3,13:15),],layout=L.3,nNodes=30,directed=FALSE,
+ edge.labels=TRUE,esize=14)
>
> qgraph(dat.3,layout=L.3,directed=FALSE,edge.labels=TRUE,esize=14,maximum=1)
> title("by Volkan OBAN using R-qgraph package",line=2.5)

explodingboxplotR package
> library(explodingboxplotR)
>
> # use this to replicate
> # from ?boxplot
> #boxplot(count ~ spray, data = InsectSprays, col = "lightgray")
>
> exploding_boxplot(
+ data.frame(
+ rowname = rownames(InsectSprays),
+ InsectSprays,
+ stringsAsFactors = FALSE),
+ y = "count",
+ group = "spray",
+ color = "spray",
+ label = "rowname"
+ )
threejs
z <- seq(-10, 10, 0.1)
x <- cos(z)
y <- sin(z)
scatterplot3js(x, y, z, color=rainbow(length(z)))
threejs
N <- 100
i <- sample(3, N, replace=TRUE)
x <- matrix(rnorm(N*3),ncol=3)
lab <- c("small", "bigger", "biggest")
scatterplot3js(x, color=rainbow(N), labels=lab[i], size=i, renderer="canvas")

threejs
data(flights)
# Approximate locations as factors
dest <- factor(sprintf("%.2f:%.2f",flights[,3], flights[,4]))
# A table of destination frequencies
freq <- sort(table(dest), decreasing=TRUE)
# The most frequent destinations in these data, possibly hub airports?
frequent_destinations <- names(freq)[1:10]
# Subset the flight data by destination frequency
idx <- dest %in% frequent_destinations
frequent_flights <- flights[idx, ]
# Lat/long and counts of frequent flights
ll <- unique(frequent_flights[,3:4])
# Plot frequent destinations as bars, and the flights to and from
# them as arcs. Adjust arc width and color by frequency.
globejs(lat=ll[,1], long=ll[,2], arcs=frequent_flights,
arcsHeight=0.3, arcsLwd=2, arcsColor="#ffff00", arcsOpacity=0.15,
atmosphere=TRUE, color="#00aaff", pointsize=0.5)

threejs
library(rgdal)
library(threejs)
# Download MODIS 16-day 1 degree Vegetation Index data manually from
# http://neo.sci.gsfc.nasa.gov/view.php?datasetId=MOD13A2_M_NDVI
# or use the following cached copy from May 25, 2014
cache <- tempfile()
writeBin(
readBin(
url("http://illposed.net/nycr2015/MOD13A2_E_NDVI_2014-05-25_rgb_360x180.TIFF",
open="rb"),
what="raw", n=1e6),
con=cache)
x <- readGDAL(cache)
# Obtain lat/long coordinates and model values as a data.frame
x <- as.data.frame(cbind(coordinates(x), x@data[,1]))
names(x) <- c("long","lat","value")
# Remove ocean areas and NA values
x <- x[x$value < 255,]
x <- na.exclude(x)
# Cut the values up into levels corresponding to the
# 99th, 95th, 90th, percentiles and then all the rest.
x$q <- as.numeric(
cut(x$value,
breaks=quantile(x$value, probs=c(0,0.90,0.95,0.99,1)),
include.lowest=TRUE))
# Colors for each level
col = c("#0055ff","#00aaff","#00ffaa","#aaff00")[x$q]
# bling out the data
globejs(lat=x$lat, long=x$long,
val=x$q^3, # Bar height
color=col,
pointsize=0.5,
atmosphere=TRUE)

DiagrammeR
library(DiagrammeR)
>
> create_random_graph(140, 100, set_seed = 23) %>%
+ join_node_attrs(get_w_connected_cmpts(.)) %>%
+ select_nodes_by_id(get_articulation_points(.)) %>%
+ set_node_attrs_ws("peripheries", 2) %>%
+ set_node_attrs_ws("width", 0.65) %>%
+ set_node_attrs_ws("height", 0.65) %>%
+ set_node_attrs_ws("penwidth", 3) %>%
+ clear_selection() %>%
+ add_global_graph_attrs(
+ attr =
+ c("color", "penwidth", "width", "height"),
+ value =
+ c("gray80", "3", "0.5", "0.5"),
+ attr_type =
+ c("edge", "edge", "node", "node")) %>%
+ colorize_node_attrs(
+ node_attr_from = "wc_component",
+ node_attr_to = "fillcolor",
+ alpha = 80) %>%
+ set_node_attr_to_display() %>%
+ select_nodes_by_degree("deg >= 3") %>%
+ trav_both_edge() %>%
+ set_edge_attrs_ws("penwidth", 4) %>%
+ set_edge_attrs_ws("color", "gray60") %>%
+ clear_selection() %>%
+ render_graph()
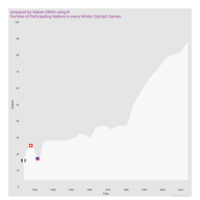
highcharter package.
ref. and code: https://www.r-bloggers.com/giving-a-thematic-touch-to-your-interactive-chart/
highcharter package.theme
ref:
https://www.r-bloggers.com/giving-a-thematic-touch-to-your-interactive-chart/

spnet package
data(world.map.simplified, package = "spnet")
graph.map.plot.position(world.map.simplified)
graph.map.plot.position(world.map.simplified, cex = 0.4)
graph.map.plot.position(world.map.simplified, label = 'ID ', cex = 0.3)

ndtv
ref: https://cran.r-project.org/web/packages/ndtv/ndtv.pdf
ndtv
data(McFarland_cls33_10_16_96)
coords<-plot(cls33_10_16_96)
# center layout coords with 100 unit area
layout.center(coords,xlim=c(0,100),ylim=c(0,100))
# rescale layout coords to unit interval
layout.normalize(coords)
qgraph
ref:https://cran.r-project.org/web/packages/qgraph/qgraph.pdf

tsna
library(networkDynamicData)
data(vanDeBunt_students)
times<-get.change.times(vanDeBunt_students)
vanDProj<-timeProjectedNetwork(vanDeBunt_students,onsets = times,termini = times)
# plot it with gray for the time edges
plot(vanDProj,
arrowhead.cex = 0,
edge.col=ifelse(vanDProj%e%'edge.type'=='within_slice','black','gray'),vertex.cex=0.7,mode='kamadakawai')
geomnet
library(geomnet)
> library(dplyr)
> # create plot
> ggplot(data = soccernet, aes(from_id = home, to_id = away)) +
+ geom_net(aes(colour = div, group = div), ealpha = .25,
+ layout.alg = 'fruchtermanreingold') +
+ facet_wrap(~season) +
+ theme_net()

geomnet
> ggplot(data = lesmisnet, aes(from_id = from, to_id = to,
+ linewidth = degree / 5 + 0.1 )) +
+ geom_net(aes(size = degree, alpha = degree),
+ colour = "grey30", ecolour = "grey60",
+ layout.alg = "fruchtermanreingold", labelon = TRUE, vjust = -0.75) +
+ scale_alpha(range = c(0.3, 1)) + theme_net() + ggtitle("by Volkan OBAN using R - geomnet")

geomnet
data(football)
ftnet <- fortify(as.edgedf(football$edges), football$vertices)
p <- ggplot(data=ftnet, aes(from_id=from_id, to_id=to_id))
p + geom_net(aes(colour=value), linewidth=0.75, size=4.5, ecolour="grey80") +
scale_colour_brewer("Conference", palette="Paired") + theme_net() +
theme(legend.position="bottom")

geomnet
emailnet <- fortify(emailedges, email$nodes, group = "day")
Joining edge and node information by from_id and label respectively.
> ggplot(data = emailnet, aes(from_id = from, to_id = to_id)) +
+ geom_net(aes(colour= CurrentEmploymentType), linewidth=0.5, fiteach=TRUE) +
+ scale_colour_brewer(palette="Set2") + facet_wrap(~day, nrow=2) + theme(legend.position="bottom") + ggtitle("by Volkan OBAN using R - geomnet")
geomnet
emailedges <- as.edgedf(subset(email$edges, nrecipients < 54))
emailnet <- fortify(emailedges, email$nodes)
#no facets
ggplot(data = emailnet, aes(from_id = from_id, to_id = to_id)) +
geom_net(aes(colour= CurrentEmploymentType), linewidth=0.5) +
scale_colour_brewer(palette="Set2")

geomnet
data(theme_elements)
TEnet <- fortify(as.edgedf(theme_elements$edges[,c(2,1)]), theme_elements$vertices)
ggplot(data = TEnet, aes(from_id = from_id, to_id = to_id)) +
geom_net(labelon=TRUE, vjust=-0.5)

geomnet
library(geomnet)
Zorunlu paket yükleniyor: ggplot2
> data(blood)
> p <- ggplot(data = blood$edges, aes(from_id = from, to_id = to))
> p + geom_net(vertices=blood$vertices, aes(colour=..type..)) + theme_net()
>
> bloodnet <- fortify(as.edgedf(blood$edges), blood$vertices)
Using from as the from node column and to as the to node column.
If this is not correct, rewrite dat so that the first 2 columns are from and to node, respectively.
Joining edge and node information by from_id and label respectively.
> p <- ggplot(data = bloodnet, aes(from_id = from_id, to_id = to_id))
> p + geom_net()
> p + geom_net(aes(colour=rho)) + theme_net()
> p + geom_net(aes(colour=rho), labelon=TRUE, vjust = -0.5)
> p + geom_net(aes(colour=rho, linetype = group_to, label = from_id),
+ vjust=-0.5, labelcolour="black", directed=TRUE) +
+ theme_net() + ggtitle(" prepared by VOLKAN OBAN using R \n geomnet package")
> p + geom_net(colour = "orange", layout.alg = 'circle', size = 6)
> p + geom_net(colour = "orange", layout.alg = 'circle', size = 6, linewidth=.75)
> p + geom_net(colour = "orange", layout.alg = 'circle', size = 0, linewidth=.75,directed = TRUE)
> p + geom_net(aes(size=Predominance, colour=rho, shape=rho, linetype=group_to),linewidth=0.75, labelon =TRUE, labelcolour="black") +
+ facet_wrap(~Ethnicity) +
+ scale_colour_brewer(palette="Set2")
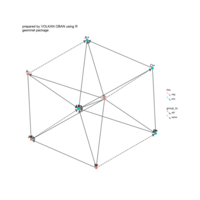
geomnet
library(geomnet)
data(blood)
p <- ggplot(data = blood$edges, aes(from_id = from, to_id = to))
p + geom_net(vertices=blood$vertices, aes(colour=..type..)) + theme_net()
bloodnet <- fortify(as.edgedf(blood$edges), blood$vertices)
p <- ggplot(data = bloodnet, aes(from_id = from_id, to_id = to_id))
p + geom_net()
p + geom_net(aes(colour=rho)) + theme_net()
p + geom_net(aes(colour=rho), labelon=TRUE, vjust = -0.5)
p + geom_net(aes(colour=rho, linetype = group_to, label = from_id),
vjust=-0.5, labelcolour="black", directed=TRUE) +
theme_net()
Latin Square
latinSq(20)
ref:http://rstudio-pubs-static.s3.amazonaws.com/1915_bd5807659c42432a9929af403b2bda5c.html

Latin Square
require(reshape2)
## Loading required package: reshape2
require(ggplot2)
## Loading required package: ggplot2
require(RColorBrewer)
## Loading required package: RColorBrewer
latinSq = function(n) {
v = rep(NA, n^2)
v[n * (1:n) - (n - 1)] = 1:n
mem = 1
for (i in 1:(n^2)) {
if (!is.na(v[i]))
mem = ifelse(v[i] < n, v[i] + 1, 1)
if (is.na(v[i])) {
v[i] = mem
mem = ifelse(mem < n, mem + 1, 1)
}
}
dim(v) = c(n, n)
lsqm = melt(v)
if (n != 7)
gg <- ggplot(lsqm, aes(x = Var1, y = Var2, fill = value, label = LETTERS[value]))
if (n == 7) {
LATINSQ = c("L", "A", "T", "I", "N", "S", "Q")[lsqm$value]
lsqm = data.frame(lsqm, LATINSQ)
gg <- ggplot(lsqm, aes(x = Var1, y = Var2, fill = value, label = LATINSQ))
}
ggPrint <- gg + geom_tile() + geom_text() + scale_fill_gradientn(colours = brewer.pal(n,
"Spectral")) + theme_bw() + theme(axis.text.x = element_blank(), axis.text.y = element_blank(),
axis.title.x = element_blank(), axis.title.y = element_blank())
ggPrint
}
latinSq(6)
languageR package
data(oldFrench)
oldFrench.ca = corres.fnc(oldFrench)
oldFrench.ca
summary(oldFrench.ca, head = TRUE)
plot(oldFrench.ca)
# more readable plot
data(oldFrenchMeta)
plot(oldFrench.ca, rlabels = oldFrenchMeta$Genre,
rcol = as.numeric(oldFrenchMeta$Genre), rcex = 0.5,
extreme = 0.1, ccol = "blue")
# create subset of proze texts
prose = oldFrench[oldFrenchMeta$Genre=="prose" &
!is.na(oldFrenchMeta$Year),]
proseinfo = oldFrenchMeta[oldFrenchMeta$Genre=="prose" &
!is.na(oldFrenchMeta$Year),]
proseinfo$Period = as.factor(proseinfo$Year <= 1250)
prose.ca = corres.fnc(prose)
plot(prose.ca, addcol = FALSE,
rcol = as.numeric(proseinfo$Period) + 1,
rlabels = proseinfo$Year, rcex = 0.7)
# and add supplementary data for texts with unknown date of composition
proseSup = oldFrench[oldFrenchMeta$Genre == "prose" &
is.na(oldFrenchMeta$Year),]
corsup.fnc(prose.ca, bycol = FALSE, supp = proseSup, font = 2,
cex = 0.8, labels = substr(rownames(proseSup), 1, 4))
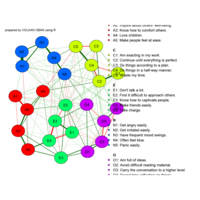
Network Graph
library("psych")
library("qgraph")
# Load BFI data:
data(bfi)
bfi <- bfi[, 1:25]
# Groups and names object (not needed really, but make the plots easier to
# interpret):
Names <- scan("http://sachaepskamp.com/files/BFIitems.txt", what = "character", sep = "\n")
# Create groups object:
Groups <- rep(c("A", "C", "E", "N", "O"), each = 5)
# Compute correlations:
cor_bfi <- cor_auto(bfi)
# Plot correlation network:
graph_cor <- qgraph(cor_bfi, layout = "spring", nodeNames = Names, groups = Groups, legend.cex = 0.6,
DoNotPlot = TRUE)
# Plot partial correlation network:
graph_pcor <- qgraph(cor_bfi, graph = "concentration", layout = "spring", nodeNames = Names,
groups = Groups, legend.cex = 0.6, DoNotPlot = TRUE)
# Plot glasso network:
graph_glas <- qgraph(cor_bfi, graph = "glasso", sampleSize = nrow(bfi), layout = "spring",
nodeNames = Names, legend.cex = 0.6, groups = Groups, legend.cex = 0.7, GLratio = 2)

meta-metafor packages
library(meta)
library(metafor)
UT_CT <- structure(list(HedgesG = c(0.423967347, 0.463106494, 0.24028285, 0.859968212,
0.700832432, -0.47267567, 1.478756303, -0.0956, 0.3216, 0.246, -0.276444701, -0.0888,
-0.0883, 0.507049057, 0.2715, 0.4705, 0.3825, 0.172067039, -0.503812571, -0.373979221,
0.268963583, 0.338268088, 0.179899652, -0.559086162, -0.0901, 0.0688, -0.211118367,
1.212322358, 0.575640797, -0.345344262, 0.929063226, 0.997507389, -0.205137778, -0.25576051,
-0.498009871, -0.330754639, 0.624634361, 0.667445161, 0.626010596, 0.03, 0.089677431,
0.30608501, -0.365244026, -0.051468156, 0.27, 0.355, 0.775529648, 1.041749533, -0.096,
-0.143722066, 0.0953, -0.5481, 0.865, -0.738, -0.3701, -0.6209, 0.2206, 0, 0.43,
-0.008883176), SE = c(0.328686052, 0.26286584, 0.204602057, 0.333714062, 0.380311448,
0.250787154, 0.40690344, 0.155084096, 0.223830293, 0.156204994, 0.319656905, 0.318168825,
0.318166748, 0.315652397, 0.214242853, 0.221133444, 0.237907545, 0.293797292, 0.301387511,
0.261597221, 0.249257982, 0.328900502, 0.233733134, 0.206587525, 0.35614549, 0.200541797,
0.171667711, 0.269412515, 0.288276271, 0.292372285, 0.33215153, 0.293760287, 0.336350481,
0.211909603, 0.23109561, 0.247283673, 0.306012425, 0.257261725, 0.326419813, 0.316,
0.247090732, 0.248441017, 0.280785825, 0.355341625, 0.2749, 0.27, 0.289786359, 0.402131319,
0.160312195, 0.157579079, 0.32046, 0.450998, 0.6359, 0.476, 0.1857, 0.2022, 0.302,
0.2455, 0.3162, 0.100200227), InverseSE = c(3.042416897, 3.804221963, 4.887536399,
2.996577349, 2.629423875, 3.987445069, 2.457585512, 6.448114433, 4.467670516, 6.401843997,
3.128354129, 3.142985494, 3.143006003, 3.168041834, 4.66760028, 4.522156316, 4.203313517,
3.403707343, 3.317987517, 3.822670572, 4.011907632, 3.040433186, 4.278383572, 4.840563347,
2.807841257, 4.986491677, 5.825207274, 3.711780056, 3.468894601, 3.420296833, 3.010674074,
3.404136109, 2.973089248, 4.718993315, 4.327213305, 4.04393864, 3.267841172, 3.887092026,
3.063539526, 3.164556962, 4.047096352, 4.025100251, 3.561433345, 2.814193243, 3.637686431,
3.703703704, 3.450818054, 2.486749858, 6.237828616, 6.346020071, 3.120514261, 2.217304733,
1.572574304, 2.100840336, 5.385029618, 4.945598417, 3.311258278, 4.073319756, 3.162555345,
9.980017326), Ap = c(1L, 1L, 1L, 1L, 1L, 0L, 1L, 0L, 1L, 1L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 1L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 1L, 0L, 0L, 1L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L), Blocked = c(0L, 1L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L, 0L,
0L, 1L, 1L, 0L, 0L, 0L, 0L, 1L, 0L, 1L, 0L, 0L, 1L, 1L, 1L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 1L, 1L, 999L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 0L, 0L, 0L, 1L, 1L, 1L, 1L, 1L, 999L,
0L, 1L, 0L, 1L, 1L), Complexity = c(48L, 60L, 36L, 48L, 48L, 48L, 48L, 48L, 48L,
30L, 48L, 48L, 48L, 48L, 48L, 48L, 48L, 40L, 40L, 48L, 48L, 60L, 48L, 48L, 48L, 48L,
48L, 108L, 108L, 36L, 48L, 48L, 48L, 48L, 48L, 48L, 48L, 48L, 160L, 48L, 48L, 36L,
44L, 48L, 144L, 144L, 48L, 36L, 48L, 40L, 48L, 48L, 48L, 75L, 48L, 48L, 96L, 48L,
48L, 48L), PresTime = c(4, 999, 2.5, 8, 8, 5, 4.5, 6, 4, 4, 8, 2, 999, 8, 8, 999,
999, 4, 999, 4, 8, 4, 8, 4, 8.8, 8.8, 999, 999, 999, 3.5, 7, 2.5, 2.5, 8, 8, 8, 10,
14, 999, 999, 999, 999, 999, 999, 4, 4, 4, 999, 4, 999, 4, 4, 4, 4, 999, 4, 999,
8, 4, 4), DelDur = c(3L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 3L, 3L, 4L, 4L,
4L, 4L, 4L, 8L, 4L, 4L, 4L, 4L, 3L, 4L, 4L, 3L, 3L, 3L, 4L, 5L, 4L, 4L, 4L, 4L, 4L,
3L, 3L, 5L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 3L, 999L, 4L, 3L, 3L, 3L, 3L, 3L, 5L, 3L,
3L, 4L, 3L, 3L), DistTask = c(3L, 3L, 1L, 1L, 1L, 2L, 2L, 1L, 2L, 2L, 4L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 3L, 1L, 3L, 1L, 1L, 1L, 1L, 1L, 3L, 3L, 1L, 1L, 1L,
1L, 1L, 4L, 4L, 1L, 4L, 4L, 4L, 4L, 4L, 1L, 1L, 1L, 1L, 4L, 1L, 2L, 2L, 2L, 2L, 1L,
1L, 3L, 1L, 1L, 2L)), .Names = c("HedgesG", "SE", "InverseSE", "Ap", "Blocked", "Complexity",
"PresTime", "DelDur", "DistTask"), class = "data.frame", row.names = c(NA, -60L))
# Code for Trim and Fill procedure, to fill in missing effect sizes.
tf1 <- trimfill(UT_CT$HedgesG, UT_CT$SE)
op <- par(cex.main = 1.5, mar = c(5, 6, 4, 5) + 0.1, mgp = c(3.5, 1, 0), cex.lab = 1.5,
font.lab = 2, cex.axis = 1.3, bty = "n", las = 1)
funnel(tf1, yaxis = "invse", xlab = "", ylab = "", contour = 0.95, xlim = c(-2, 2), ylim = c(1,
12), cex = 2, col = "black", col.contour = "lightgray", ref = 0, axes = F)
axis(1)
axis(2)
par(las = 0)
mtext("Hedges' G", side = 1, line = 2.5, cex = 1.5)
mtext("Inverse of Standard Error", side = 2, line = 3, cex = 1.5)
par(op)

Questionnaire Graph
library("psych")
library("qgraph")
# Load BFI data:
data(bfi)
bfi <- bfi[, 1:25]
# Groups and names object (not needed really, but make the plots easier to
# interpret):
Names <- scan("http://sachaepskamp.com/files/BFIitems.txt", what = "character", sep = "\n")
# Create groups object:
Groups <- rep(c("A", "C", "E", "N", "O"), each = 5)
# Compute correlations:
cor_bfi <- cor_auto(bfi)
# Plot correlation network:
graph_cor <- qgraph(cor_bfi, layout = "spring", nodeNames = Names, groups = Groups, legend.cex = 0.6,
DoNotPlot = TRUE)
# Plot partial correlation network:
graph_pcor <- qgraph(cor_bfi, graph = "concentration", layout = "spring", nodeNames = Names,
groups = Groups, legend.cex = 0.6, DoNotPlot = TRUE)
# Plot glasso network:
graph_glas <- qgraph(cor_bfi, graph = "glasso", sampleSize = nrow(bfi), layout = "spring",
nodeNames = Names, legend.cex = 0.6, groups = Groups, legend.cex = 0.7, GLratio = 2,
DoNotPlot = TRUE)
# centrality plot (all graphs):
centralityPlot(list(r = graph_cor, `Partial r` = graph_pcor, glasso = graph_glas),
labels = Names) + labs(colour = "") + theme_bw() + theme(legend.position = "bottom")

Plot
> FacVar1 = as.factor(rep(c("level1", "level2"), 25))
> FacVar2 = as.factor(rep(c("levelA", "levelB", "levelC"), 17)[-51])
> FacVar3 = as.factor(rep(c("levelI", "levelII", "levelIII", "levelIV"), 13)[-c(51:52)])
>
> ## 4 Numeric Vars
> set.seed(123)
> NumVar1 = round(rnorm(n = 50, mean = 1000, sd = 50), digits = 2) ## Normal distribution
> set.seed(123)
> NumVar2 = round(runif(n = 50, min = 500, max = 1500), digits = 2) ## Uniform distribution
> set.seed(123)
> NumVar3 = round(rexp(n = 50, rate = 0.001)) ## Exponential distribution
> NumVar4 = 2001:2050
>
> simData = data.frame(FacVar1, FacVar2, FacVar3, NumVar1, NumVar2, NumVar3, NumVar4)
> plot(simData$NumVar1,main="by VOLKAN OBAN using R", type = "o", ylim = c(0, max(simData$NumVar1, simData$NumVar2))) ## index plot with one variable
> lines(simData$NumVar2, type = "o", lty = 2, col = "purple")
>
streamgraph in R.
library(streamgraph)
> library(viridis)
>
> stocks_url <- "http://infographics.economist.com/2015/tech_stocks/data/stocks.csv"
> stocks <- read.csv(stocks_url, stringsAsFactors=FALSE)
>
> stock_colors <- viridis_pal()(100)
> stocks %>%
+ mutate(date=as.Date(quarter, format="%m/%d/%y")) %>%
+ streamgraph(key="ticker", value="nominal", offset="expand") %>%
+ sg_fill_manual(stock_colors) %>%
+ sg_axis_x(tick_interval=10, tick_units="year") %>%
+ sg_legend(TRUE, "Ticker: ")

ggmap
ref: https://mran.microsoft.com/web/packages/ggmap/ggmap.pdf
corrr package-Correlations in R
mtcars
A tool for exploring correlations.
It makes it possible to easily perform routine tasks when
exploring correlation matrices such as ignoring the diagonal,
focusing on the correlations of certain variables against others,
or rearranging and visualising the matrix in terms of the
strength of the correlations

ggraph
require(igraph)
gr <- graph_from_data_frame(flare$edges, vertices = flare$vertices)
ggraph(gr, 'treemap', weight = 'size') + geom_node_tile()
# We can color by modifying the graph
gr <- tree_apply(gr, function(node, parent, depth, tree) {
if (depth == 1) {
tree <- set_vertex_attr(tree, 'Class', node, V(tree)$shortName[node])
} else if (depth > 1) {
tree <- set_vertex_attr(tree, 'Class', node, V(tree)$Class[parent])
}
tree
})
ggraph
> require(igraph)
> flareGraph <- graph_from_data_frame(flare$edges, vertices = flare$vertices)
> ggraph(flareGraph, 'dendrogram', circular = TRUE) +
+ geom_edge_diagonal0() +
+ geom_node_text(aes(filter = leaf, angle = node_angle(x, y), label = shortName),
+ hjust = 'outward', size = 2) +
+ expand_limits(x = c(-1.3, 1.3), y = c(-1.3, 1.3))
> require(igraph)
> flareGraph <- graph_from_data_frame(flare$edges, vertices = flare$vertices)
> ggraph(flareGraph, 'dendrogram', circular = TRUE) +
+ geom_edge_diagonal0() +
+ geom_node_text(aes(filter = leaf, angle = node_angle(x, y), label = shortName),
+ hjust = 'outward', size = 2) +
+ expand_limits(x = c(-1.3, 1.3), y = c(-1.3, 1.3))

ggforce ggraph
> library(igraph)
> graph <- graph_from_data_frame(highschool)
> ggraph(graph) + geom_edge_link() + geom_node_point() + theme_graph()
Using `nicely` as default layout
>
> library(ggforce)
> sizes <- sample(10, 100, TRUE)
> position <- pack_circles(sizes)
> data <- data.frame(x = position[,1], y = position[,2], r = sqrt(sizes/pi))
> ggplot() +
+ geom_circle(aes(x0 = x, y0 = y, r = r), data = data, fill = 'steelblue') +
+ geom_circle(aes(x0 = 0, y0 = 0, r = attr(position, 'enclosing_radius'))) +
+ geom_polygon(aes(x = x, y = y),
+ data = data[attr(position, 'front_chain'), ],
+ fill = NA,
+ colour = 'black')

ggraph
require(igraph)
gr <- graph_from_data_frame(flare$edges, vertices = flare$vertices)
ggraph(gr, 'treemap', weight = 'size') + geom_node_tile()
# We can color by modifying the graph
gr <- tree_apply(gr, function(node, parent, depth, tree) {
if (depth == 1) {
tree <- set_vertex_attr(tree, 'Class', node, V(tree)$shortName[node])
} else if (depth > 1) {
tree <- set_vertex_attr(tree, 'Class', node, V(tree)$Class[parent])
}
tree
})
ggraph(gr, 'treemap', weight = 'size') +
geom_node_tile(aes(fill = Class, filter = leaf, alpha = depth), colour = NA) +
geom_node_tile(aes(size = depth), colour = 'white') +
scale_alpha(range = c(1, 0.5), guide = 'none') +
scale_size(range = c(4, 0.2), guide = 'none')

ggraph
> require(igraph)
> gr <- graph_from_data_frame(flare$edges, vertices = flare$vertices)
> ggraph(gr, 'circlepack', weight = 'size') + geom_node_circle() + coord_fixed()

ggraph
> library(igraph)
> gr <- graph_from_data_frame(highschool)
> V(gr)$popularity <- as.character(cut(degree(gr, mode = 'in'), breaks = 3,
+ labels = c('low', 'medium', 'high')))
> ggraph(gr) +
+ geom_edge_link() + geom_node_point() +
+ facet_nodes(~popularity)

ggraph
> gr <- graph_from_data_frame(highschool)
> ggraph(gr) +
+ geom_edge_link() +
+ geom_node_point() +
+ facet_edges(~year)
Using `nicely` as default layout
>
> library(igraph)
> gr <- graph_from_data_frame(highschool)
> ggraph(gr) +
+ geom_edge_link() +
+ geom_node_point() +
+ facet_edges(~year)

Plot
variety=c(rep("soldur" , 40), rep("silur" , 40), rep("lloyd" , 40),
rep("pescadou" , 40) , rep("X4582" , 40) ,
rep("Dudur" , 40) , rep("Classic" , 40))
treatment= rep(c(rep("high" , 20) , rep("low" , 20)) , 7)
note=c( rep(c(sample(0:4, 20 , replace=T) , sample(1:6, 20 , replace=T)),2),
rep(c(sample(5:7, 20 , replace=T), sample(5:9, 20 , replace=T)),2),
c(sample(0:4, 20 , replace=T) , sample(2:5, 20 , replace=T),
rep(c(sample(6:8, 20 , replace=T) , sample(7:10, 20 , replace=T)),2) ))
data=data.frame(variety, treatment , note)
new_order <- with(data, reorder(variety , note, mean , na.rm=T))
# Then I make the boxplot, asking to use the 2 factors : variety (in the good order) AND treatment :
par(mar=c(3,4,3,1))
myplot=boxplot(note ~ treatment*new_order , data=data , boxwex=0.4 , ylab="sickness",
main="sickness of several wheat lines" , col=c("slateblue1" , "tomato") , xaxt="n")
# To add the label of x axis
my_names=sapply(strsplit(myplot$names , '\\.') , function(x) x[[2]] )
my_names=my_names[seq(1 , length(my_names) , 2)]
axis(1, at = seq(1.5 , 14 , 2), labels = my_names , tick=FALSE , cex=0.3)
for(i in seq(0.5 , 20 , 2)){ abline(v=i,lty=1, col="grey")}
# Add a legend
legend("bottomright", legend = c("High treatment", "Low treatment"), col=c("slateblue1" , "tomato"),
pch = 15, bty = "n", pt.cex = 3, cex = 1.2, horiz = F, inset = c(0.1, 0.1))

Plot
m <- matrix(c(1,1,1,
2,3,4,
5,6,7), ncol=3, by=T)
l <- layout(m)
layout.show(l) # show layout to doublecheck
# layout cells are filled in the order of the numbers
# set par, e.g. mar each time if required
for (i in 1:7) {
par(mar=c(i,i,i,i))
hist(rnorm(100), col=i)
}
multigraph
bmgraph(swomen, layout = "bip3", cex = 3, tcex = .8, pch = c(19, 15), lwd = 1.5, vcol = 2:3)
ref:https://github.com/mplex/multigraph

multigraph
> swomen <- read.dl(file = "http://moreno.ss.uci.edu/davis.dat")
> bmgraph(swomen,main="\n prepared by Volkan OBAN using R \n multigraph package

multigraph
floflies <- read.dl(file = "http://moreno.ss.uci.edu/padgett.dat")
multigraph(floflies, directed = FALSE, layout = "force", seed = 2, cex = 6, tcex = .7, pos = 0, vcol = 8,ecol = 1, lwd = 2, bwd = .5, lty = 2:1, pch = 13)

ggplot2 and ggthemes
theme_calc()

ggplot2 and ggthemes
ref:https://www.r-bloggers.com/how-to-create-a-data-visualization-from-the-new-york-times-in-r/
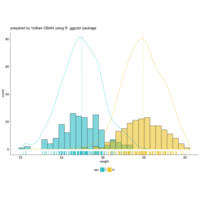
ggpubr
set.seed(1234)
wdata = data.frame(
sex = factor(rep(c("F", "M"), each=200)),
weight = c(rnorm(200, 55), rnorm(200, 58)))
head(wdata, 4)
gghistogram(wdata, x = "weight",
add = "mean", rug = TRUE,
fill = "sex", palette = c("#00AFBB", "#E7B800"),
add_density = TRUE)

ggpubr
data("ToothGrowth")
df <- ToothGrowth
ggdotplot(df, "dose", "len",
add = "boxplot",
color = "dose", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"))
ggpubr
ggdotchart(df, x = "mpg", main="by VOLKAN OBAN", label = "name",
group = "cyl", color = "cyl",palette = "Dark2" )

ggpubr
data("mtcars")
df <- mtcars
df$cyl <- as.factor(df$cyl)
df$name <- rownames(df)
head(df[, c("wt", "mpg", "cyl")], 3)
# Basic plot
ggdotchart(df, x = "mpg", label = "name" )
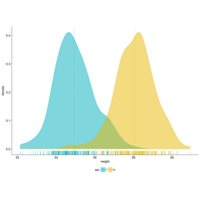
ggpubr
wdata = data.frame(
+ sex = factor(rep(c("F", "M"), each=200)),
+ weight = c(rnorm(200, 55), rnorm(200, 58)))
> head(wdata, 4)
sex weight
1 F 53.79293
2 F 55.27743
3 F 56.08444
4 F 52.65430
>
> ggdensity(wdata, x = "weight", fill = "lightgray",
+ add = "mean", rug = TRUE)
> ggdensity(wdata, x = "weight",
+ add = "mean", rug = TRUE,
+ color = "sex", fill = "sex",
+ palette = c("#00AFBB", "#E7B800"))

ggpubr
df <- ToothGrowth
ggboxplot(df, "dose", "len",
fill = "dose", palette = c("#00AFBB", "#E7B800", "#FC4E07"))

ggpubr
> data("ToothGrowth")
> df <- ToothGrowth
ggboxplot(df, x = "dose", y = "len",
add = "jitter", shape = "dose")
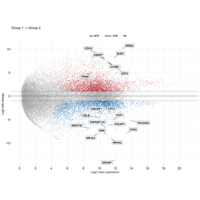
ggpubr
data(diff_express)
ggmaplot(diff_express, main = expression("Group 1" %->% "Group 2"),
fdr = 0.05, fc = 2, size = 0.4,
palette = c("#B31B21", "#1465AC", "darkgray"),
genenames = as.vector(diff_express$name),
legend = "top", top = 20,
font.label = c("bold", 11), label.rectangle = TRUE,
font.legend = "bold",
font.main = "bold",
ggtheme = ggplot2::theme_minimal())

ggpubr
ggviolin(df, x = "dose", y = "len", fill = "dose",
palette = c("#00AFBB", "#E7B800", "#FC4E07"),
add = "boxplot", add.params = list(fill = "white"))

ggplot2
sp <- ggplot(faithful, aes(x=eruptions, y=waiting)) +
geom_point()
sp + geom_density_2d()
# Gradient de couleur
sp + stat_density_2d(aes(fill = ..level..), geom="polygon")
# Changer le gradient de couleur
sp + stat_density_2d(aes(fill = ..level..), geom="polygon")+
scale_fill_gradient(low="blue", high="red")

ggplot2
sp <- ggplot(faithful, aes(x=eruptions, y=waiting)) +
geom_point()
sp + geom_density_2d()
# Gradient de couleur
sp + stat_density_2d(aes(fill = ..level..), geom="polygon")
# Changer le gradient de couleur
sp + stat_density_2d(aes(fill = ..level..), geom="polygon")+
scale_fill_gradient(low="blue", high="red")

horzintal boxplot
library(ggplot2)
d <- diamonds
levels(d$cut) <- list(A="Fair", B="Good", " "="space1", C="Very Good", D="Ideal", " "="space2", E="Premium")
ggplot(d, aes(x=cut, y=depth)) +
geom_boxplot(color="black", size=0.2) +
theme_bw() +
scale_x_discrete(breaks = c("A", "B", " ", "C", "D", " ", "E"), drop=FALSE) +
coord_flip()
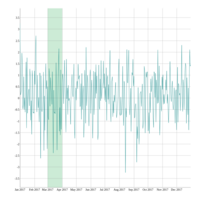
manipulateWidget
if (require(dygraphs) && require(xts)) {
mydata <- xts(rnorm(365), order.by = as.Date("2017-01-01") + 0:364)
manipulateWidget(
dygraph(mydata) %>% dyShading(from=period[1], to = period[2], color = "#CCEBD6"),
period = mwDateRange(c("2017-03-01", "2017-04-01"),
min = "2017-01-01", max = "2017-12-31")
)
}
manipulateWidget
ref:https://cran.rstudio.com/web/packages/manipulateWidget/manipulateWidget.pdf

WVplots package
set.seed(34903490)
x = rnorm(50)
y = 0.5*x^2 + 2*x + rnorm(length(x))
frm = data.frame(x=x,y=y,yC=y>=as.numeric(quantile(y,probs=0.8)))
frm$absY <- abs(frm$y)
frm$posY = frm$y > 0
frm$costX = 1
WVPlots::DoubleHistogramPlot(frm, "x", "yC", title="Example double histogram plot")

BatchGetSymbols package
library(BatchGetSymbols)
first.date <- Sys.Date()-150
last.date <- Sys.Date()
tickers <- c('FB','NYSE:MMM','PETR4.SA','abcdef')
l.out <- BatchGetSymbols(tickers = tickers,
first.date = first.date,
last.date = last.date)
library(ggplot2)
p <- ggplot(l.out$df.tickers, aes(x = ref.date, y = price.close))
p <- p + geom_line()
p <- p + facet_wrap(~ticker, scales = 'free_y')
print(p)

stick package
require(stick)
> set.seed(68331)
> plotStick(x = runif(100), y = runif(100))

edgebundleR
> require(igraph)
> ws_graph <- watts.strogatz.game(1, 50, 4, 0.05)
> edgebundle(ws_graph,tension = 0.1,fontsize = 18,padding=40)

edgebundleR
ref: https://github.com/garthtarr/edgebundleR

edgebundleR
require(huge)
data("stockdata")
# generate returns sequences
X = log(stockdata$data[2:1258,]/stockdata$data[1:1257,])
# perform some regularisation
out.huge = huge(cor(X),method = "glasso",lambda=0.56,verbose = FALSE)
# identify the linkages
adj.mat = as.matrix(out.huge$path[[1]])
# format the colnames
nodenames = paste(gsub("","",stockdata$info[,2]),stockdata$info[,1],sep=".")
head(cbind(stockdata$info[,2],stockdata$info[,1],nodenames))
colnames(adj.mat) = rownames(adj.mat) = nodenames
# restrict attention to the connected stocks:
adj.mat = adj.mat[rowSums(adj.mat)>0,colSums(adj.mat)>0]
# plot the result
edgebundle(adj.mat,tension=0.8,fontsize = 10)

timevis
> data <- data.frame(
+ id = 1:4,
+ content = c("geldim" , "gördüm" ,"dünya", "gideceğim"),
+ start = c("1984-01-24", "2010-01-11", "2020-12-20", "2016-02-14 15:00:00"),
+ end = c(NA,NA, "2016-02-04", NA))
>
> timevis(data)
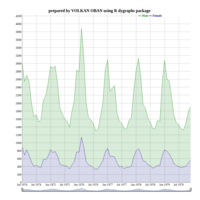
dygraphs
> dygraph(lungDeaths)
> dygraph(lungDeaths,main = "prepared by VOLKAN OBAN using R \n dygraphs package") %>%
+ dySeries("mdeaths", label = "Male") %>%
+ dySeries("fdeaths", label = "Female") %>%
+ dyOptions(stackedGraph = TRUE) %>%
+ dyRangeSelector(height = 20)

ggplot2
library(ggplot2)
# Create a Violin plot
ggplot(diamonds, aes(x = cut, y = price, fill = clarity)) +
geom_violin(trim= FALSE) + scale_y_log10() +
facet_wrap(~ clarity)
ggplot2
ggplot(diamonds, aes(x = cut, y = price, fill = clarity)) +
geom_violin() + scale_y_log10()

ggplot2
> ggplot(diamonds, aes(x = cut, y = price, fill = cut)) +
+ geom_violin() + scale_y_log10() +
+ geom_boxplot(width = 0.2)

ggplot2
gplot(ChickWeight, aes(x = Diet, y = weight, color = Diet)) +
+ geom_violin(fill = "pink") +
+ geom_jitter(position = position_jitter(0.2)) +
+ theme(legend.position = "top")

ggplot2
> ggplot(ChickWeight, aes(x = Diet, y = weight)) +
geom_boxplot(notch = TRUE) +
geom_jitter(position = position_jitter(0.5), aes(colour = Diet)

stripchart
> data(airquality)
> # prepare the data
> temp <- airquality$Temp
>
> # gererate normal distribution with same mean and sd
> tempNorm <- rnorm(200,mean=mean(temp, na.rm=TRUE), sd = sd(temp, na.rm=TRUE))
>
> # make a list
> x <- list("temp"=temp, "norm"=tempNorm)
> stripchart(x,
+ main="prepared by VOLKAN OBAN using R \n Multiple stripchart for comparision",
+ xlab="Degree Fahrenheit",
+ ylab="Temperature",
+ method="jitter",
+ col=c("purple","red"),
+ pch=16
+ )

ggplot2
p <- ggplot(mpg, aes(cyl, hwy))
p + geom_point()
p + geom_jitter()
p + geom_jitter(aes(colour = class))

lattice package-stripchart
df = data.frame(y = rnorm(500), x = sample(LETTERS[1:5],500,replace=T)
library(lattice)
boxplot(y ~ x, data = ddf, lwd = 2,xlab='x', ylab = 'y')
stripchart(y ~ x, vertical = TRUE, data = ddf,method = "jitter", add = TRUE, pch = 20, col = 'purple')

lattice package
ref:https://science.nature.nps.gov/im/datamgmt/statistics/r/advanced/latticegraphics.cfm

qplot
> year <- function(x) as.POSIXlt(x)$year + 1900
> qplot(unemploy / pop, uempmed, data = economics,
+ geom = c("point", "path")) + ggtitle("prepared by VOLKAN OBAN using R-ggplot2 - data(economics) ")

ggplot2
qplot(color, price / carat, data = diamonds, geom = "jitter",alpha = I(1 / 5) )

ggplot2
> library(arules)
> data("AdultUCI")
> dframe = AdultUCI[, c("education", "hours-per-week")]
> colnames(dframe) = c("education", "hours_per_week")
> library(ggplot2)
> ggplot(dframe, aes(x=education, y=hours_per_week)) +
+ geom_point(colour="lightblue", alpha=0.1, position="jitter") +
+ geom_boxplot(outlier.size=0.5, alpha=0.2) + coord_flip()
qrage package
library(qrage)
> data(links)
> #Data that determines the color of the nodes
> data(nodeColor)
> #Data that determines the size of the node
> data(nodeValues)
> #Create graph
> qrage(links=links,nodeColor=nodeColor,nodeValue=nodeValues,cut=0.1)
>
rpivotTable
library(dplyr)
iris %>%
tbl_df %>%
filter( Sepal.Width > 3 ) %>%
rpivotTable

sjPlot and sjmisc package
> library(sjmisc)
> data(efc)
> sjp.grpfrq(efc$e17age, efc$e16sex, show.values = FALSE)
>
> sjp.grpfrq(efc$e17age, efc$e42dep, intr.var = efc$e16sex, type = "box")

higncharter package
> data(worldgeojson, package = "highcharter")
> data("GNI2014", package = "treemap")
> highchart(type = "map") %>%
+ hc_add_series_map(map = worldgeojson, df = GNI2014, value = "GNI", joinBy = "iso3") %>%
+ hc_colorAxis(stops = color_stops()) %>%
+ hc_tooltip(useHTML = TRUE, headerFormat = "",
+ pointFormat = "this is {point.name} and have {point.population} people with gni of {point.GNI}")

higncharter package
> hciconarray(c("nice", "good"), c(10, 20))
> hciconarray(c("nice", "good"), c(10, 20), size = 10)
> hciconarray(c("nice", "good"), c(100, 200), icons = "child")
> hciconarray(c("car", "truck", "plane"), c(75, 30, 20), icons = c("car", "truck", "plane")) %>%
+ hc_add_theme(
+ hc_theme_merge(
+ hc_theme_flatdark(),
+ hc_theme_null(chart = list(backgroundColor = "#34495e"))
+ )
+ )
higncharter package
> hciconarray(c("nice", "good"), c(10, 20))
> hciconarray(c("nice", "good"), c(10, 20), size = 10)
> hciconarray(c("nice", "good"), c(100, 200), icons = "child")
> hciconarray(c("car", "truck", "plane"), c(75, 30, 20), icons = c("car", "truck", "plane")) %>%
+ hc_add_theme(
+ hc_theme_merge(
+ hc_theme_flatdark(),
+ hc_theme_null(chart = list(backgroundColor = "#34495e"))
+ )
+ )

ggplot2
http://r-statistics.co/Top50-Ggplot2-Visualizations-MasterList-R-Code.html#Marginal%20Histogram%20/%20Boxplot

ggplot2
library(ggplot2)
theme_set(theme_bw())
# plot
g <- ggplot(mpg, aes(manufacturer, cty))
g + geom_boxplot() +
geom_dotplot(binaxis='y',
stackdir='center',
dotsize = .5,
fill="red") +
theme(axis.text.x = element_text(angle=65, vjust=0.6)) +
labs(title="Box plot + Dot plot",
subtitle="City Mileage vs Class: Each dot represents 1 row in source data",
caption="Source: mpg",
x="Class of Vehicle",
y="City Mileage")

Plot
df = structure(list(Affiliation = structure(c(1L, 2L, 3L, 4L,
5L, 6L, 7L, 8L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 1L, 2L, 3L, 4L,
5L, 6L, 7L, 8L), .Label = c("BMI", "CCS",
"CS", "Epi", "Genom", "HSE",
"HSR", "HPR"), class = "factor"),
count = structure(c(4L, 21L, 14L, 20L, 11L, 13L, 19L, 15L,
5L, 22L, 17L, 24L, 9L, 12L, 18L, 16L, 1L, 10L, 7L, 23L, 2L,
3L, 8L, 6L), .Label = c("15", "26", "27", "32", "40", "41",
"42", "58", "62", "63", "70", "88", "89", "96", "99", "112",
"125", "160", "164", "172", "176", "178", "200", "628"), class = "factor"),
Year = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L), .Label = c("2014",
"2015", "2016"), class = "factor")), .Names = c("Affiliation",
"count", "Year"), row.names = c(NA, 24L), class = "data.frame")
ggplot(df, aes(x = Affiliation, y = count, fill = Year, group = Year)) +
geom_bar(position = position_dodge(width = 0.9), stat = "identity", alpha = 1,
size = 1, width = 0.05) +
geom_text(aes(label = count), position = position_dodge(width = 0.9),
vjust = -0.25) + scale_fill_brewer(palette = "Set1")
df$count <- as.numeric(as.character(df$count))
gg <- ggplot(df, aes(Affiliation, count))
gg <- gg + geom_segment(aes(xend=Affiliation, yend=0))
gg <- gg + geom_point()
gg <- gg + geom_text(aes(label=count, y=count+25), vjust=0, size=3)
gg <- gg + scale_y_continuous(expand=c(0,0), limits=c(0, 800))
gg <- gg + facet_wrap(~Year)
gg <- gg + labs(x=NULL, y=NULL)
gg <- gg + theme_bw()
gg <- gg + theme(strip.background=element_blank())
gg <- gg + theme(strip.text=element_text(hjust=0))
gg <- gg + theme(panel.grid.major.x=element_blank())
gg <- gg + theme(panel.grid.minor.y=element_blank())
gg <- gg + theme(axis.ticks=element_blank())
gg <- gg + theme(axis.text.x=element_text(size=8))
gg <- gg + theme(axis.text.y=element_text(size=8, vjust=c(0, 0.5, 0.5, 0.5, 1)))
gg

caroline package
n <- rnorm(130, 10, 3)
p <- rpois(110, 4)
u <- runif(300, 0, 20)
l <- rlnorm(130, log(2))
g <- rgamma(140, 3)
e <- rexp(160)
violins(list(e=e, p=p,u=u,n=n,l=l,g=g), ylim=c(0,20),
col=c('purple','lightblue','lightgreen','red','orange','yellow'),
stats=TRUE)

stripchart
> ds = read.csv("http://www.math.smith.edu/r/data/help.csv")
> smallds = subset(ds, female==1)
> boxplot(pcs~homeless, data=smallds,
+ horizontal=TRUE)
> stripchart(round(pcs)~homeless,
+ method='stack', data=smallds,
+ add=TRUE) + title("by VOLKAN OBAN")

DescTools
library(DescTools)
library(Sleuth3)
attach(ex0923)
PlotBubble( x= Educ, y = AFQT, area = Income2005/1000,
col = SetAlpha(as.numeric(Gender)), border = "burlywood",
inches = .5, xlab = "Education", ylab = "AFQT test score")
title(main = "Income, proportional to circle area")
legend("left", c("Female","Male"),
text.col = c(1:2), cex =.9, bty = "n")

corrplot
> library(corrplot)
> library(Sleuth2)
> attach(ex1713)
>y = cor(ex1713[, 2:6])
> par(mfrow = c(2,2))
> corrplot(y) # default method is "circle"
> corrplot(y, method = "color")
> corrplot(y, method = "number")
> corrplot(y, method = "ellipse", type = "lower"

corrgram
> library(corrgram)
> col.corrgram <- function(ncol){
+ colorRampPalette(c("darkgoldenrod4", "burlywood1",
+ "darkkhaki", "darkgreen"))(ncol)}
> corrgram(mtcars, order=TRUE, lower.panel=panel.shade,
+ upper.panel=panel.pie, text.panel=panel.txt,
+ main="prepared by Volkan OBAN using R-corrgram \n Correlogram of Car Mileage Data (PC2/PC1 Order)")

corrgram
> corrgram(mtcars, order=TRUE, lower.panel=panel.ellipse,
+ upper.panel=panel.pts, text.panel=panel.txt,
+ diag.panel=panel.minmax,
+ main="prepared by Volkan OBAN using R-corrgram \n Car Milage Data in PC2/PC1 Order")

plotrix
library(plotrix)
#Build the matrix data to look like a correlation matrix
n <- 8
x <- matrix(runif(n*n), nrow=n)
xmin <- 0
xmax <- 1
for (i in 1:n) x[i, i] <- 1.0 #Make the diagonal all 1's
#Generate the palette for the matrix and the legend. Generate labels for the legend
palmat <- color.scale(x, c(1, 0.4), c(1, 0.4), c(0.96, 1))
palleg <- color.gradient(c(1, 0.4), c(1, 0.4), c(0.96, 1), nslices=100)
lableg <- c(formatC(xmin, format="f", digits=2), formatC(1*(xmax-xmin)/4, format="f", digits=2), formatC(2*(xmax-xmin)/4, format="f", digits=2), formatC(3*(xmax-xmin)/4, format="f", digits=2), formatC(xmax, format="f", digits=2))
#Set up the plot area and plot the matrix
par(mar=c(5, 5, 5, 8))
color2D.matplot(x, cellcolors=palmat, main=paste(n, " X ", n, " Matrix Using Color2D.matplot", sep=""), show.values=2, vcol=rgb(0,0,0), axes=FALSE, vcex=0.7)
axis(1, at=seq(1, n, 1)-0.5, labels=seq(1, n, 1), tck=-0.01, padj=-1)
#In the axis() statement below, note that the labels are decreasing. This is because
#the above color2D.matplot() statement has "axes=FALSE" and a normal axis()
#statement was used.
axis(2, at=seq(1, n, 1)-0.5, labels=seq(n, 1, -1), tck=-0.01, padj=0.7)
#Plot the legend
pardat <- par()
color.legend(pardat$usr[2]+0.5, 0, pardat$usr[2]+1, pardat$usr[2], paste(" ", lableg, sep=""), palleg, align="rb", gradient="y", cex=0.7)

gplots
> library(gplots)
>
> #Build the matrix data to look like a correlation matrix
> x <- matrix(rnorm(64), nrow=8)
> x <- (x - min(x))/(max(x) - min(x)) #Scale the data to be between 0 and 1
> for (i in 1:8) x[i, i] <- 1.0 #Make the diagonal all 1's
>
> #Format the data for the plot
> xval <- formatC(x, format="f", digits=2)
> pal <- colorRampPalette(c(rgb(0.96,0.96,1), rgb(0.1,0.1,0.9)), space = "rgb")
>
> #Plot the matrix
> x_hm <- heatmap.2(x, Rowv=FALSE, Colv=FALSE, dendrogram="none", main="8 X 8 Matrix Using Heatmap.2", xlab="using R-gplots", ylab="", col=pal, tracecol="#303030", trace="none", cellnote=xval, notecol="black", notecex=0.8, keysize = 1.5, margins=c(5, 5))
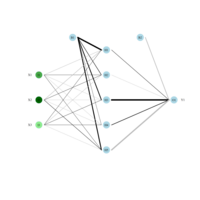
Plotnet
library(neuralnet)
mod <- neuralnet(Y1 ~ X1 + X2 + X3, data = neuraldat, hidden = 5)
plotnet(mod,main="by Volkan OBAN")
## using caret
library(caret)
mod <- train(Y1 ~ X1 + X2 + X3, method = 'nnet', data = neuraldat, linout = TRUE)
plotnet(mod)
## a more complicated network with categorical response
AND <- c(rep(0, 7), 1)
OR <- c(0, rep(1, 7))
binary_data <- data.frame(expand.grid(c(0, 1), c(0, 1), c(0, 1)), AND, OR)
mod <- neuralnet(AND + OR ~ Var1 + Var2 + Var3, binary_data,
hidden = c(6, 12, 8), rep = 10, err.fct = 'ce', linear.output = FALSE)
plotnet(mod,main="by Volkan OBAN")
## recreate the previous example with numeric inputs
# get the weights and structure in the right format
wts <- neuralweights(mod)
struct <- wts$struct
wts <- unlist(wts$wts)
# plot
plotnet(wts, struct = struct,main="by Volkan OBAN")
## color input nodes by relative importance
mod <- nnet(Y1 ~ X1 + X2 + X3, data = neuraldat, size = 5)
rel_imp <- garson(mod, bar_plot = FALSE)$rel_imp
cols <- colorRampPalette(c('lightgreen', 'darkgreen'))(3)[rank(rel_imp)]
plotnet(mod, circle_col = list(cols, 'lightblue'),main="by Volkan OBAN")

lattice package in R
bwplot(gcsescore ~ gender | factor(score), Chem97, layout = c(6, 1))

lattice package in R
data(Chem97, package = "mlmRev")
bwplot(factor(score) ~ gcsescore | gender, Chem97)
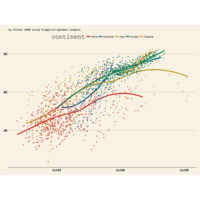
ggplot2
ref: http://bioconnector.org/bims8382/r-ggplot2.html

ggplot2
ref:http://bioconnector.org/bims8382/r-ggplot2.html

Multinomial Logistic Regression
ref:http://www.ats.ucla.edu/stat/r/dae/mlogit.htm

hcpc hierarchical clustering on principal components hybrid approach
http://www.sthda.com/english/wiki/hcpc-hierarchical-clustering-on-principal-components-hybrid-approach-2-2-unsupervised-machine-learning

Visualize kmeans clustering
> set.seed(123)
> # K-means clustering
> km.res <- kmeans(scale(USArrests), 4, nstart = 25)
> # Use clusplot function
> library(cluster)
> clusplot(scale(USArrests), km.res$cluster, main = "Cluster plot",
+ color=TRUE, labels = 2, lines = 0)
> library("factoextra")
> # Visualize kmeans clustering
> fviz_cluster(km.res, USArrests)

ggplot2
> library(ggplot2)
> library(dplyr)
> data(diamonds)
> diamonds %>%
+ ggplot(aes(x=carat,y=price)) +
+ geom_point(alpha=0.5) +
+ facet_grid(~ cut) +
+ stat_smooth(method = lm, formula = y ~ poly(x,2)) +
+ theme_bw()

CatterPlots
library(CatterPlots)
meow <- multicat(xs=x, ys=rnorm(21),
cat=c(1,2,3,4,5,6,7,8,9,10),
catcolor=list(c(0,0,0,1)),
canvas=c(-0.1,1.1, -0.1, 1.1),
xlab="some cats", ylab="other cats", main="Random Cats")
ref:https://github.com/Gibbsdavidl/CatterPlots

GGally
> ggpairs(iris, upper=list(continuous="density"),
lower=list(continuous="smooth",combo="facetdensity"), color="Species")

GGally
> require(GGally)
> ggpairs(iris, color='Species', alpha=0.4)

ggplot2
dist <- data.frame(value=rnorm(10000, 1:4), group=1:4)
myBoxplot + scale_fill_discrete(breaks=c("1","3","2","4"), labels=c("Dist 1","Dist
3","Dist 2","Dist 4"))
myBoxplot + theme_bw()
ggplot2
aa <- data.frame(value=rnorm(dist <- data.frame(value=rnorm(10000, 1:4), group=1:4)10000, 1:4), group=1:4)
ggplot(aa, aes(x=group, y=value, color=group)) + geom_jitter(alpha=0.5)

ggplot2
https://rpubs.com/ikochergin/177292
ggplot2
dist <- data.frame(value=rnorm(10000, 1:4), group=1:4)
ggplot(dist, aes(x=group, y=value, color=group)) +
geom_jitter(alpha=0.2,shape=21)

ggplot2
ggplot(data=myMovieData,
aes(Type,Budget)) +
geom_jitter() +
geom_boxplot(alpha=I(0.6))+
scale_y_log10()

ggplot2
library(ggplot2movies)
glimpse(movies)
d1 <-data.frame(movies[movies$Action==1, c("budget", "Short",
"year")])
d1$Type <- "Animation"
d2 <-data.frame(movies[movies$Animation==1, c("budget", "Short",
"year")])
d2$Type <- "Animation"
d3 <-data.frame(movies[movies$Comedy==1, c("budget", "Short",
"year")])
d3$Type <- "Comedy"
d4 <-data.frame(movies[movies$Drama==1, c("budget", "Short",
"year")])
d4$Type <- "Drama"
d5 <-data.frame(movies[movies$Documentary==1, c("budget", "Short",
"year")])
d5$Type <- "Documentary"
d6 <-data.frame(movies[movies$Romance==1, c("budget", "Short",
"year")])
d6$Type <- "Romance"
myMovieData <- rbind(d1, d2, d3, d4, d5, d6)
names(myMovieData) <- c("Budget", "Short", "Year", "Type" )
glimpse(myMovieData)
qplot(Type,Budget, data=myMovieData, geom=c("boxplot","jitter"), log="y")

ggplot2 and maps packages
data(world.cities)
capitals <- subset(world.cities, capital == 1)
capitals.big <- subset(capitals, pop > 3000000)
ggplot(capitals.big, aes(long, lat)) + borders("world") + geom_point(aes(size =
pop)) + geom_text(aes(long, lat,label=country.etc),hjust=-0.2,size=4)
ggplot(capitals.big, aes(long, lat)) + borders("world") + geom_point(aes(size =
pop)) + geom_text(aes(long, lat,label=country.etc),hjust=-0.2,size=4) +
coord_map(projection = "ortho", orientation=c(41, 20, 0))
ggplot2 and maps packages
> data(world.cities)
> capitals <- subset(world.cities, capital == 1)
> ggplot(capitals, aes(long, lat)) + borders("world", fill="cornflowerblue",
col="darkorchid") + geom_point(aes(size = pop),col="blueviolet")

ggplot2 and gridExtra packages.
p1 <- ggplot(diamonds, aes(x=carat, y=price, color=cut)) + geom_point() + geom_smooth() + theme(legend.position="none") + labs(title="legend.position='none'") # remove legend
p2 <- ggplot(diamonds, aes(x=carat, y=price, color=cut)) + geom_point() + geom_smooth() + theme(legend.position="top") + labs(title="legend.position='top'") # legend at top
p3 <- ggplot(diamonds, aes(x=carat, y=price, color=cut)) + geom_point() + geom_smooth() + labs(title="legend.position='coords inside plot'") + theme(legend.justification=c(1,0), legend.position=c(1,0)) # legend inside the plot.
grid.arrange(p1, p2, p3, ncol=3)

ggplot2 facet_wrap
ref:
http://sharpsightlabs.com/blog/small-multiples-ggplot/

ggplot2
library(ggplot2)
library(grid)
library(reshape2)
options(stringsAsFactors=FALSE)
# Generating synthetic data here
tpl <- c('1st', '2nd', '3rd', '4th', '5th')
dat <- data.frame(foo=as.factor(sample(tpl, 1000, replace=TRUE)),
bar=as.factor(sample(tpl, 1000, replace=TRUE)),
effect=runif(1000, 0.1, 0.7))
# Just doing a cross-tabulation
ctab <- melt(table(subset(dat, select=c('foo', 'bar'))), id.vars='foo')
ctab$y <- rep(0.8, dim(ctab)[1])
# Just conducting ANOVA tests here
tests <- c()
for (q in levels(dat$bar)) {
test <- aov(effect ~ foo, data=subset(dat, bar == q))
tests <- c(tests, sprintf('p-value: ~%.4f', summary(test)[[1]][['Pr(>F)']][[1]]))
}
tests <- data.frame(p.value=tests, bar=levels(dat$bar),
x=rep(1, 5), y=rep(0, 5))
ggplot(dat, mapping=aes(y=effect)) +
geom_boxplot(mapping=aes(x=foo)) +
geom_text(data=tests, aes(x=x, y=y, label=p.value), hjust=0.1, vjust=0.1) +
geom_text(data=ctab, aes(x=foo, y=y, label=value), vjust=0.7) +
xlab('2000 Census White Pop. Proportion Quintile') +
ylab('Vegetation Cover Proportion') +
labs(title='Vegetation Cover by 2000 Census Tract, Pop. Density Quintiles') +
facet_wrap(~ bar) +
theme_bw() +
theme(text=element_text(size=16),
plot.margin=unit(c(0.5, 0.2, 0.5, 0), 'cm'),
panel.grid.major.y=element_line(color='gray'),
panel.grid.major.x=element_blank())

ggplot2 facet_wrap
ggplot(mpg, aes(displ, hwy)) +
geom_point(data = transform(mpg, class = NULL), colour = "grey85") +
geom_point() +
facet_wrap(~class)
ggplot2 facetwrap
p <- qplot(price, data = diamonds, geom = "histogram", binwidth = 1000) + ggtitle("by Volkan OBAN using R - ggplot2")
> p + facet_wrap(~ color)
> p + facet_wrap(~ color, scales = "free_y")
> p <- qplot(displ, hwy, data = mpg)
> p + facet_wrap(~ cyl)
> p + facet_wrap(~ cyl, scales = "free")
> cyl6 <- subset(mpg, cyl == 6)
> p + geom_point(data = cyl6, colour = "red", size = 1) +
+ facet_wrap(~ cyl)
> p + geom_point(data = transform(cyl6, cyl = 7), colour = "red") +
+ facet_wrap(~ cyl)
> p + geom_point(data = transform(cyl6, cyl = NULL), colour = "red") +
+ facet_wrap(~ cyl)
>

ggplot2 facetwrap
d <- ggplot(diamonds, aes(carat, price, fill = ..density..)) +
xlim(0, 2) + stat_binhex(na.rm = TRUE) + opts(aspect.ratio = 1)
d + facet_wrap(~ color)
d + facet_wrap(~ color, ncol = 1)
d + facet_wrap(~ color, ncol = 4)
d + facet_wrap(~ color, nrow = 1)
d + facet_wrap(~ color, nrow = 3)
# Using multiple variables continues to wrap the long ribbon of
# plots into 2d - the ribbon just gets longer
# d + facet_wrap(~ color + cut)
ggplot2
https://www.ling.upenn.edu/~joseff/rstudy/summer2010_ggplot2_intro.html

ggplot2
ggplot(mpg, aes(class, hwy, fill = factor(year)))+
+ geom_boxplot()
> ggplot(mpg, aes(reorder(class, hwy, median), hwy, fill = factor(year)))+
geom_boxplot()

ggplot2
> p <- ggplot(mpg, aes(displ, hwy))
>
> p + geom_point() + stat_smooth()
`geom_smooth()` using method = 'loess'
> p + geom_point() + stat_smooth(method = "lm")
>
> library(MASS)
Attaching package: ‘MASS’
The following object is masked from ‘package:plotly’:
select
The following object is masked from ‘package:dplyr’:
select
> p + geom_point() + stat_smooth(method = "rlm")
> p + stat_smooth(geom = "point")+stat_smooth(geom = "errorbar")

ggalt
d <- data.frame(x=c(1,1,2),y=c(1,2,2)*100)
> gg <- ggplot(d,aes(x,y))
> gg <- ggplot(mpg, aes(displ, hwy))
> gg + geom_encircle(data=subset(mpg, hwy>40)) + geom_point()
> gg + geom_encircle(aes(group=manufacturer)) + geom_point()
> gg + geom_encircle(aes(group=manufacturer,fill=manufacturer),alpha=0.4)+
+ geom_point()
> gg + geom_encircle(aes(group=manufacturer,fill=manufacturer),alpha=0.4)+
+ geom_point()
Social Network Analysis with R using Package igraph
library(igraph)
ref:https://rdatamining.wordpress.com/2012/05/17/an-example-of-social-network-analysis-with-r-using-package-igraph/
rbokeh
> p <- figure(width = 1000) %>%
+ ly_points(date, Freq, data = flightfreq,
+ hover = list(date, Freq, dow), size = 5) %>%
+ ly_abline(v = as.Date("2001-09-11"))
> p
>

rbokeh
> figure(data = lattice::singer) %>%
+ ly_points(catjitter(voice.part), jitter(height), color = "black") %>%
+ ly_boxplot(voice.part, height, with_outliers = FALSE)

rbokeh
idx <- split(1:150, iris$Species)
figs <- lapply(idx, function(x) {
figure(width = 300, height = 300) %>%
ly_points(Sepal.Length, Sepal.Width, data = iris[x, ],
hover = list(Sepal.Length, Sepal.Width))
})
# 1 row, 3 columns
grid_plot(figs)
# specify xlim and ylim to be applied to all panels
grid_plot(figs, xlim = c(4, 8), ylim = c(1.5, 4.5))
# unnamed list will remove labels
grid_plot(unname(figs))
# 2 rows, 2 columns
grid_plot(figs, nrow = 2)
# x and y axis with same (and linked) limits
grid_plot(figs, same_axes = TRUE)
# x axis with same (and linked) limits
grid_plot(figs, same_axes = c(TRUE, FALSE), nrow = 2)
# x axis with same (and linked) limits and custom xlim
grid_plot(figs, same_axes = c(TRUE, FALSE), xlim = c(5, 7), nrow = 2)
# send lists instead of specifying nrow and ncol
grid_plot(list(
c(list(figs[[1]]), list(figs[[3]])),
c(list(NULL), list(figs[[2]]))
))
# a null entry will be skipped in the grid
figs2 <- figs
figs2[1] <- list(NULL)
grid_plot(figs2, nrow = 2)
# with themes
grid_plot(figs) %>%
theme_title(text_color = "red") %>%
theme_plot(background_fill_color = "#E6E6E6",
outline_line_color = "white") %>%
theme_grid(c("x", "y"), grid_line_color = "white",
minor_grid_line_color = "white",
minor_grid_line_alpha = 0.4) %>%
theme_axis(c("x", "y"), axis_line_color = "white",
major_label_text_color = "#7F7F7F",
major_tick_line_color = "#7F7F7F",
minor_tick_line_alpha = 0, num_minor_ticks = 2)
# themes again
grid_plot(figs) %>%
set_theme(bk_ggplot_theme)
# link data across plots in the grid (try box_select tool)
# (data sources must be the same)
tools <- c("pan", "wheel_zoom", "box_zoom", "box_select", "reset")
p1 <- figure(tools = tools, width = 500, height = 500) %>%
ly_points(Sepal.Length, Sepal.Width, data = iris, color = Species)
p2 <- figure(tools = tools, width = 500, height = 500) %>%
ly_points(Petal.Length, Petal.Width, data = iris, color = Species)
grid_plot(list(p1, p2), same_axes = TRUE, link_data = TRUE)
circlize package
> circos.clear()
> layout(matrix(1:9, 3, 3))
> for(i in 1:9) {
+ factors = 1:8
+ par(mar = c(0.5, 0.5, 0.5, 0.5))
+ circos.par(cell.padding = c(0, 0, 0, 0))
+ circos.initialize(factors, xlim = c(0, 1))
+ circos.trackPlotRegion(ylim = c(0, 1), track.height = 0.05,
+ bg.col = rand_color(8), bg.border = NA)
+ for(i in 1:20) {
+ se = sample(1:8, 2)
+ circos.link(se[1], runif(2), se[2], runif(2),
+ col = rand_color(1, transparency = 0.4), border = NA)
+ }
+ circos.clear()
+ }

heatmaply
> library(heatmaply)
> heatmaply(iris[,-5], k_row = 3, k_col = 2,main="by VOLKAN OBAN using R \n heatmaply package-data(iris)")
> heatmaply(cor(iris[,-5]))
> heatmaply(cor(iris[,-5]), limits = c(-1,1))
> heatmaply(mtcars, k_row = 3, k_col = 2)

ggplot2
library(ggplot2)
> library(reshape)
> data(HairEyeColor)
> P=t(HairEyeColor[,,2])
> Pm=melt(P)
ggplot(Pm, aes(Eye, Hair, fill=value)) + geom_tile() +
geom_text(aes(label=Pm$value),colour="white")+
theme(axis.text.x=element_text(size = 15),axis.text.y=element_text(size = 15))

ggTimeSeries
library(ggplot2)
library(ggthemes)
library(data.table)
library(ggTimeSeries)
set.seed(1)
dtData = data.table(
DateCol = seq(
as.Date("1/01/2014", "%d/%m/%Y"),
as.Date("31/12/2015", "%d/%m/%Y"),
"days"
),
ValueCol = runif(730)
)
dtData[, ValueCol := ValueCol + (strftime(DateCol,"%u") %in% c(6,7) * runif(1) * 0.75), .I]
dtData[, ValueCol := ValueCol + (abs(as.numeric(strftime(DateCol,"%m")) - 6.5)) * runif(1) * 0.75, .I]
dtData[, CategCol := letters[1 + round(ValueCol * 7)]]
# base plot
p2 = ggplot_calendar_heatmap(
dtData,
'DateCol',
'CategCol'
)
# adding some formatting
p2 +
xlab('') +
ylab('') +
facet_wrap(~Year, ncol = 1)

Plot
set.seed(1)
dfData = data.frame(x = 1:1000, y = cumsum(rnorm(1000)))
# base plot
p1 = ggplot_horizon(dfData, 'x', 'y')
p1 +
xlab('') +
ylab('') +
scale_fill_continuous(low = 'green', high = 'red') +
coord_fixed( 0.5 * diff(range(dfData$x)) / diff(range(dfData$y)))
Waterfall
set.seed(1)
dfData = data.frame(x = 1:100, y = cumsum(rnorm(100)))
# base plot
p1 = ggplot_waterfall(
dtData = dfData,
'x',
'y'
)
# adding some formatting
p1 +
xlab('') +
ylab('')

plotKML
ref:https://cran.r-project.org/web/packages/plotKML/plotKML.pdf
data(eberg)
data(eberg_grid)
data(eberg_zones)
data(eberg_contours)
library(sp)
coordinates(eberg) <- ~X+Y
proj4string(eberg) <- CRS("+init=epsg:31467")
gridded(eberg_grid) <- ~x+y
proj4string(eberg_grid) <- CRS("+init=epsg:31467")
# visualize the maps:
data(SAGA_pal)
l.sp <- list("sp.lines", eberg_contours, col="black")
## Not run:
spplot(eberg_grid["DEMSRT6"], col.regions = SAGA_pal[[1]], sp.layout=l.sp)
spplot(eberg_zones, sp.layout=list("sp.points", eberg, col="black", pch="+"))
ggplot2
library(ggplot2)
library(reshape)
require(PerformanceAnalytics)
data(edhec)
ed=data.frame(edhec)
ed$date=as.Date(rownames(ed))
m=melt(ed,id="date")
m$variable=gsub('\\.',' ',m$variable)
ggplot(m,aes(date,0,fill=value))+geom_tile(aes(height=max(m$value)-min(m$value)))+geom_line(aes(x=date,y=value))+facet_grid(variable~.)+ scale_fill_gradient2(low="red",high="blue")+ylab("value") +xlab("Date \n by VOLKAN OBAN using R")
kmeans
df=iris
> m=as.matrix(cbind(df$Petal.Length, df$Petal.Width),ncol=2)
> cl=(kmeans(m,3))
> df$cluster=factor(cl$cluster)
> centers=as.data.frame(cl$centers)
> library(ggplot2)
>
> ggplot(data=df, aes(x=Petal.Length, y=Petal.Width, color=cluster )) +
+ geom_point() +
+ geom_point(data=centers, aes(x=V1,y=V2, color='Center')) +
+ geom_point(data=centers, aes(x=V1,y=V2, color='Center'), size=52, alpha=.3,)

barplot
> data(BreastCancer)
> # create a bar plot of each categorical attribute
> par(mfrow=c(2,4))
> for(i in 2:9) {
+ counts <- table(BreastCancer[,i])
+ name <- names(BreastCancer)[i]
+ barplot(counts, main=name)
+ }
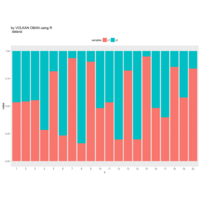
Plot
> library(ggplot2)
> library(dplyr)
> library(tidyr)
>
> dfr <- data.frame(x=factor(1:20),y1=runif(n=20)) %>%
+ mutate(y2=1-y1) %>%
+ gather(variable,value,-x)
ggplot(dfr,aes(x=x,y=value,fill=variable))+
geom_bar(stat="identity")+
labs(title=" title")+
theme(legend.position="top",
legend.justification="right")

R Data viz.
ref: http://timelyportfolio.blogspot.com.tr/
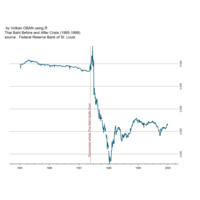
R Data viz.
ref: http://timelyportfolio.blogspot.com.tr/

VIM
data(tao, package = "VIM")
aggr(tao)

ComplexHeatmap
library(circlize)
library(RColorBrewer)
library(ComplexHeatmap)
lt = readRDS(paste0(system.file(package = "ComplexHeatmap"), "/extdata/meth.rds"))
list2env(lt, envir = environment())
ha = HeatmapAnnotation(df = data.frame(type = c(rep("Tumor", 10), rep("Control", 10))),
col = list(type = c("Tumor" = "red", "Control" = "blue")))
ha2 = HeatmapAnnotation(df = data.frame(type = c(rep("Tumor", 10), rep("Control", 10))),
col = list(type = c("Tumor" = "red", "Control" = "blue")), show_legend = FALSE)
# column order of the methylation matrix which will be assigned to the expressio matrix
column_tree = hclust(dist(t(meth)))
ht_list =
Heatmap(meth, name = "methylation", col = colorRamp2(c(0, 0.5, 1), c("blue", "white", "red")),
cluster_columns = column_tree, top_annotation = ha, column_names_gp = gpar(fontsize = 8), km = 5,
column_title = "Methylation", column_title_gp = gpar(fontsize = 10),
row_title_gp = gpar(fontsize = 10)) +
Heatmap(direction, name = "direction", col = c("hyper" = "red", "hypo" = "blue"),
column_names_gp = gpar(fontsize = 8)) +
Heatmap(expr[, column_tree$order], name = "expression", col = colorRamp2(c(-2, 0, 2), c("green", "white", "red")),
cluster_columns = FALSE, top_annotation = ha2, column_names_gp = gpar(fontsize = 8),
column_title = "Expression", column_title_gp = gpar(fontsize = 10)) +
Heatmap(cor_pvalue, name = "-log10(cor_p)", col = colorRamp2(c(0, 2, 4), c("white", "white", "red")),
column_names_gp = gpar(fontsize = 8)) +
Heatmap(gene_type, name = "gene type", col = brewer.pal(length(unique(gene_type)), "Set1"),
column_names_gp = gpar(fontsize = 8)) +
Heatmap(anno, name = "anno_gene", col = brewer.pal(length(unique(anno)), "Set2"),
column_names_gp = gpar(fontsize = 8)) +
Heatmap(dist, name = "dist_tss", col = colorRamp2(c(0, 10000), c("black", "white")),
column_names_gp = gpar(fontsize = 8)) +
Heatmap(enhancer, name = "anno_enhancer", col = colorRamp2(c(0, 1), c("white", "orange")),
cluster_columns = FALSE, column_names_gp = gpar(fontsize = 8), column_title = "Enhancer",
column_title_gp = gpar(fontsize = 10))
ht_global_opt(heatmap_legend_title_gp = gpar(fontsize = 8, fontface = "bold"),
heatmap_legend_labels_gp = gpar(fontsize = 8))
draw(ht_list, newpage = FALSE, column_title = "prepared by Volkan OBAN using R-ComplexHeatmap \n Correspondence between methylation, expression and other genomic features",
column_title_gp = gpar(fontsize = 12, fontface = "bold"), heatmap_legend_side = "bottom")
invisible(ht_global_opt(RESET = TRUE))

ComplexHeatmap pvclust
library(ComplexHeatmap)
library(MASS)
library(pvclust)
data(Boston)
boston.pv <- pvclust(Boston, nboot=100)
plot(boston.pv)

heatmap
mat = readRDS(paste0(system.file("extdata", package = "ComplexHeatmap"), "/measles.rds"))
ha1 = HeatmapAnnotation(dist1 = anno_barplot(colSums(mat), bar_width = 1, gp = gpar(col = NA, fill = "#FFE200"),
border = FALSE, axis = TRUE))
ha2 = rowAnnotation(dist2 = anno_barplot(rowSums(mat), bar_width = 1, gp = gpar(col = NA, fill = "#FFE200"),
border = FALSE, which = "row", axis = TRUE), width = unit(1, "cm"))
ha_column = HeatmapAnnotation(cn = function(index) {
year = as.numeric(colnames(mat))
which_decade = which(year %% 10 == 0)
grid.text(year[which_decade], which_decade/length(year), 1, just = c("center", "top"))
})
Heatmap(mat, name = "cases", col = colorRamp2(c(0, 800, 1000, 127000), c("white", "cornflowerblue", "yellow", "red")),
cluster_columns = FALSE, show_row_dend = FALSE, rect_gp = gpar(col= "white"), show_column_names = FALSE,
row_names_side = "left", row_names_gp = gpar(fontsize = 10),
column_title = 'Measles cases in US states 1930-2001\nVaccine introduced 1961',
top_annotation = ha1, top_annotation_height = unit(1, "cm"),
bottom_annotation = ha_column, bottom_annotation_height = grobHeight(textGrob("1900"))) + ha2
decorate_heatmap_body("cases", {
i = which(colnames(mat) == "1961")
x = i/ncol(mat)
grid.lines(c(x, x), c(0, 1), gp = gpar(lwd = 2))
grid.text("Vaccine introduced", x, unit(1, "npc") + unit(5, "mm"))
})

VIM
marginplot(sleep[c("Gest","Dream")],pch = c(20),col=c("purple","yellow","pink"))
R time-series forecasting with neural network-nnetar
> x<- c(1774, 1706, 1288, 1276, 2350, 1821, 1712, 1654, 1680, 1451,
+ 1275, 2140, 1747, 1749, 1770, 1797, 1485, 1299, 2330, 1822, 1627,
+ 1847, 1797, 1452, 1328, 2363, 1998, 1864, 2088, 2084, 594, 884,
+ 1968, 1858, 1640, 1823, 1938, 1490, 1312, 2312, 1937, 1617, 1643,
+ 1468, 1381, 1276, 2228, 1756, 1465, 1716, 1601, 1340, 1192, 2231,
+ 1768, 1623, 1444, 1575, 1375, 1267, 2475, 1630, 1505, 1810, 1601,
+ 1123, 1324, 2245, 1844, 1613, 1710, 1546, 1290, 1366, 2427, 1783,
+ 1588, 1505, 1398, 1226, 1321, 2299, 1047, 1735, 1633, 1508, 1323,
+ 1317, 2323, 1826, 1615, 1750, 1572, 1273, 1365, 2373, 2074, 1809,
+ 1889, 1521, 1314, 1512, 2462, 1836, 1750, 1808, 1585, 1387, 1428,
+ 2176, 1732, 1752, 1665, 1425, 1028, 1194, 2159, 1840, 1684, 1711,
+ 1653, 1360, 1422, 2328, 1798, 1723, 1827, 1499, 1289, 1476, 2219,
+ 1824, 1606, 1627, 1459, 1324, 1354, 2150, 1728, 1743, 1697, 1511,
+ 1285, 1426, 2076, 1792, 1519, 1478, 1191, 1122, 1241, 2105, 1818,
+ 1599, 1663, 1319, 1219, 1452, 2091, 1771, 1710, 2000, 1518, 1479,
+ 1586, 1848, 2113, 1648, 1542, 1220, 1299, 1452, 2290, 1944, 1701,
+ 1709, 1462, 1312, 1365, 2326, 1971, 1709, 1700, 1687, 1493, 1523,
+ 2382, 1938, 1658, 1713, 1525, 1413, 1363, 2349, 1923, 1726, 1862,
+ 1686, 1534, 1280, 2233, 1733, 1520, 1537, 1569, 1367, 1129, 2024,
+ 1645, 1510, 1469, 1533, 1281, 1212, 2099, 1769, 1684, 1842, 1654,
+ 1369, 1353, 2415, 1948, 1841, 1928, 1790, 1547, 1465, 2260, 1895,
+ 1700, 1838, 1614, 1528, 1268, 2192, 1705, 1494, 1697, 1588, 1324,
+ 1193, 2049, 1672, 1801, 1487, 1319, 1289, 1302, 2316, 1945, 1771,
+ 2027, 2053, 1639, 1372, 2198, 1692, 1546, 1809, 1787, 1360, 1182,
+ 2157, 1690, 1494, 1731, 1633, 1299, 1291, 2164, 1667, 1535, 1822,
+ 1813, 1510, 1396, 2308, 2110, 2128, 2316, 2249, 1789, 1886, 2463,
+ 2257, 2212, 2608, 2284, 2034, 1996, 2686, 2459, 2340, 2383, 2507,
+ 2304, 2740, 1869, 654, 1068, 1720, 1904, 1666, 1877, 2100, 504,
+ 1482, 1686, 1707, 1306, 1417, 2135, 1787, 1675, 1934, 1931, 1456)
> y=auto.arima(x)
> plot(forecast(y,h=30))
> points(1:length(x),fitted(y),type="l",col="green"
+ )
> library(caret)
> fit <- nnetar(x)
> plot(forecast(fit,h=60)
> points(1:length(x),fitted(fit),type="l",col="green")
>

tmap
tm_shape(World, bbox = "Turkey") +
+ tm_borders("grey20") +
+ tm_grid(projection="longlat", labels.size = .5) +
+ tm_text("name", size="AREA") +
+ tm_compass(position = c(.65, .15), color.light = "grey90") +
+ tm_credits("Eckert IV projection", position = c(.85, 0)) +
+ tm_style_classic(inner.margins=c(.04,.03, .02, .01), legend.position = c("left", "bottom"),
+ legend.frame = TRUE, bg.color="lightblue", legend.bg.color="lightblue", title="by Volkan OBAN using R- tmap \n TURKEY",
+ earth.boundary = TRUE, space.color="grey90")
>

party package
irisct <- ctree(Species ~ .,data = iris)
irisct
plot(irisct)
table(predict(irisct), iris$Species)

Multipanel Graphics in R
library(rgdal)
par(mfrow=c(1,3))
plot(Sepal.Length, Sepal.Width, col='skyblue', pch=1)
title("Sepal.Length vs Sepal.Width")
plot(Sepal.Length, Petal.Length, col='magenta', pch=2)
title("Sepal.Length vs Petal.Length")
plot(Sepal.Length, Petal.Width, col='red', pch=3)
title("Sepal.Length vs Petal.Width")

GGally package
library(GGally)
ds = read.csv("http://www.math.smith.edu/r/data/help.csv")
ds$sex = as.factor(ifelse(ds$female==1, "female", "male"))
ds$housing = as.factor(ifelse(ds$homeless==1, "homeless", "housed"))
smallds = subset(ds, select=c("housing", "sex", "i1", "cesd"))
ggpairs(smallds, diag=list(continuous="density", discrete="bar"), axisLabels="show")

ggplot2 and ggthemes
P <- ggplot(data = mpg,aes(cty, hwy,color=class))+geom_point(size=3) + facet_wrap(~ manufacturer,scales="free")+
+ labs(title=" prepared by Volkan OBAN \n data = mpg --ggplot2 and ggthemes packages \n Plot With Facets")
> P
> P +scale_colour_Publication()+ theme_Publication()
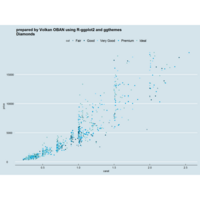
ggplot2 and ggthemes
(qplot(carat, price, data=dsamp, colour=cut)
+ theme_economist()
+ scale_colour_economist()
+ ggtitle("Diamonds Are Forever"))

ggplot2
dtemp <- data.frame(months = factor(rep(substr(month.name,1,3), 4), levels = substr(month.name,1,3)),
city = rep(c("Tokyo", "New York", "Berlin", "London"), each = 12),
temp = c(7.0, 6.9, 9.5, 14.5, 18.2, 21.5, 25.2, 26.5, 23.3, 18.3, 13.9, 9.6,
-0.2, 0.8, 5.7, 11.3, 17.0, 22.0, 24.8, 24.1, 20.1, 14.1, 8.6, 2.5,
-0.9, 0.6, 3.5, 8.4, 13.5, 17.0, 18.6, 17.9, 14.3, 9.0, 3.9, 1.0,
3.9, 4.2, 5.7, 8.5, 11.9, 15.2, 17.0, 16.6, 14.2, 10.3, 6.6, 4.8))
ggplot(dtemp, aes(x = months, y = temp, group = city, color = city)) +
geom_line() +
geom_point(size = 1.1) +
ggtitle("Monthly Average Temperature") +
theme_hc() +
scale_colour_hc()
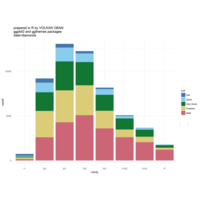
ggplot2 and ggthemes
ggplot(diamonds, aes(x = clarity, fill = cut)) +
geom_bar() +
scale_fill_ptol() +
theme_minimal()
ggmap
library(ggmap)
# example of map of Dhangadhi, Nepal
dhanmap1 = get_map(location = c(lon = 29.016896, lat = 41.118085 ,zoom = 12, maptype = 'roadmap', source = "google")
dhanmap1 = ggmap(dhanmap1)
dhanmap1

ggplot2
set.seed (78888)
rectheat = sample(c(rnorm (10, 5,1), NA, NA), 150, replace = T)
circlefill = rectheat*10 + rnorm (length (rectheat), 0, 3)
circlesize = rectheat*1.5 + rnorm (length (rectheat), 0, 3)
myd <- data.frame (rowv = rep (1:10, 15), columnv = rep(1:15, each = 10),
rectheat, circlesize, circlefill)
require(ggplot2)
pl1 <- ggplot(myd, aes(y = factor(rowv), x = factor(columnv))) + geom_tile(aes(fill = rectheat)) + scale_fill_continuous(low = "blue", high = "green")
pl1 + geom_point(aes(colour = circlefill, size =circlesize)) + scale_color_gradient(low = "yellow", high = "red")+ scale_size(range = c(1, 20))+ theme_bw()

epade package
> x<-rnorm(1000)
> g<-round(runif(1000))
> g2<-round(runif(1000))
> box.plot.ade(x, g, g2, vnames=list(c("subgroup 1","subgroup 2"),
+ c("group 1", "group 2")), wall=0, count='N: ?', means=TRUE)
> box.plot.ade(x, g, g2, vnames=list(c("subgroup 1","subgroup 2"),
+ c("group 1", "group 2")), wall=1, type="violin")
> box.plot.ade(x, g, g2, vnames=list(c("subgroup 1","subgroup 2"),
+ c("group 1", "group 2")), wall=2, type="percentile")
> box.plot.ade(x, g, g2, vnames=list(c("subgroup 1","subgroup 2"),
+ c("group 1", "group 2")), wall=3, type="sd")

epade package
> x<-rnorm(1000)
> g<-round(runif(1000))
> g2<-round(runif(1000))
> box.plot.ade(x, g, g2, vnames=list(c("subgroup 1","subgroup 2"),
+ c("group 1", "group 2")), wall=0, count='N: ?', means=TRUE)
> box.plot.ade(x, g, g2, vnames=list(c("subgroup 1","subgroup 2"),
+ c("group 1", "group 2")), wall=1, type="violin")
> box.plot.ade(x, g, g2, vnames=list(c("subgroup 1","subgroup 2"),
+ c("group 1", "group 2")), wall=2, type="percentile")
> box.plot.ade(x, g, g2, vnames=list(c("subgroup 1","subgroup 2"),
+ c("group 1", "group 2")), wall=3, type="sd")

epade package
> x<-rnorm(1000)
> g<-round(runif(1000))
> g2<-round(runif(1000))
> box.plot.ade(x, g, g2, vnames=list(c("subgroup 1","subgroup 2"),
+ c("group 1", "group 2")), wall=0, count='N: ?', means=TRUE)
> box.plot.ade(x, g, g2, vnames=list(c("subgroup 1","subgroup 2"),
+ c("group 1", "group 2")), wall=1, type="violin")
> box.plot.ade(x, g, g2, vnames=list(c("subgroup 1","subgroup 2"),
+ c("group 1", "group 2")), wall=2, type="percentile")
> box.plot.ade(x, g, g2, vnames=list(c("subgroup 1","subgroup 2"),
+ c("group 1", "group 2")), wall=3, type="sd")

epade package
x<- round(runif(1000, 0.5, 10.5))
bar.plot.ade(x, btext='Uniform distribution', gradient=TRUE)
x<-rbinom(1000, 1, 0.75)
y<-rbinom(1000, 1, 0.30)
z<-rbinom(1000, 1, 0.50)
bar.plot.ade(x,y,z)
bar.plot.ade(x,y,z, wall=4, form='c', main='Bar-Plot')
ggplot2
> ggplot(mtcars,aes(x = cyl, y = mpg)) +
geom_violin(fill = "pink") +
geom_point(aes(size = carb), colour = "blue", position = "jitter")
+ xlab("cyl") + ylab ("mpg")

"rworldmap"
> library(rworldmap)
> d <- data.frame(
+ country=c("Turkey", "Italy", "Germany", "AZERBAIJAN","SPAIN"),
+ value=c(-2, -1, 0, 1, 2))
n <- joinCountryData2Map(d, joinCode="NAME", nameJoinColumn="country")
mapCountryData(n, nameColumnToPlot="value", mapTitle="prepared by Volkan OBAN using R \n ",xlim=c(-20, 70), ylim=c(15, 70),colourPalette="red2White",addLegend=TRUE,oceanCol="lightblue", missingCountryCol="purple")

"rworldmap"
> library(rworldmap)
> d <- data.frame(
+ country=c("Turkey", "France", "Germany", "Italy", "Netherlands"),
+ value=c(-2, -1, 0, 1, 2))
> n <- joinCountryData2Map(d, joinCode="NAME", nameJoinColumn="country")
> mapCountryData(n, nameColumnToPlot="value", mapTitle="World"

ggplot2
library(ggplot2)
library(ggthemes)
ggplot(mtcars, aes(wt, mpg)) + geom_point() + geom_rug() + theme_tufte(ticks=F) +
xlab("Car weight (lb/1000)") + ylab("Miles per gallon of fuel") +
theme(axis.title.x = element_text(vjust=-0.5), axis.title.y = element_text(vjust=1))
plotrix
testlen<-c(rnorm(36)*2+5)
testpos<-seq(0,350,by=10)
polar.plot(testlen,testpos,main="Test Polar Plot",lwd=3,line.col=4)
polar.plot(testlen,testpos,main="Test Clockwise Polar Plot",
start=90,clockwise=TRUE,lwd=3,line.col=4)

plotrix
testcp<-list("",40)
for(i in 1:40) testcp[[i]]<-rnorm(sample(1:8,1)*50)
segs<-get.segs(testcp)
centipede.plot(segs,main="Test centipede plot",vgrid=0)

genoplotR
ref: http://genoplotr.r-forge.r-project.org/code/barto_seg_plots.R

feature Plot
> featurePlot(x = iris[, 1:4],
+ y = iris$Species,
+ plot = "box",
+ ## Pass in options to bwplot()
+ scales = list(y = list(relation="free"),
+ x = list(rot = 90)),
+ layout = c(4,1 ) ,main=" feature Plot",
+ auto.key = list(columns = 2))

AppliedPredictiveModeling
library(AppliedPredictiveModeling)
> transparentTheme(trans = .4)
> library(caret)
> featurePlot(x = iris[, 1:4],
+ y = iris$Species,
+ plot = "pairs",main="prepared by Volkan OBAN using R",
+ auto.key = list(columns = 3))

outbreaks
ref: https://shiring.github.io/machine_learning/2016/11/27/flu_outcome_ML_post

outbreaks
ggplot(data = fluH7N9.china.2013_gather, aes(x = Date, y = age, fill = outcome)) +
stat_density2d(aes(alpha = ..level..), geom = "polygon") +
geom_jitter(aes(color = outcome, shape = gender), size = 1.5) +
geom_rug(aes(color = outcome)) +
labs(
fill = "Outcome",
color = "Outcome",
alpha = "Level",
shape = "Gender",
x = "Date in 2013",
y = "Age",
title = "2013 Influenza A H7N9 cases in China",
subtitle = "Dataset from 'outbreaks' package (Kucharski et al. 2014)",
caption = ""
) +
facet_grid(Group ~ province) +
my_theme() +
scale_shape_manual(values = c(15, 16, 17)) +
scale_color_brewer(palette="Set1", na.value = "grey50") +
scale_fill_brewer(palette="Set1")

rpart
tree1 <- rpart(survived~., data=ptitanic)
par(mfrow=c(4,3))
for(iframe in 1:nrow(tree1$frame)) {
cols <- ifelse(1:nrow(tree1$frame) <= iframe, "black", "gray")
prp(tree1, col=cols, branch.col=cols, split.col=cols)
}

rpart
heat.tree <- function(tree, low.is.green=FALSE, ...) { # dots args passed to prp
y <- tree$frame$yval
if(low.is.green)
y <- -y
max <- max(y)
min <- min(y)
cols <- rainbow(99, end=.36)[
ifelse(y > y[1], (y-y[1]) * (99-50) / (max-y[1]) + 50,
(y-min) * (50-1) / (y[1]-min) + 1)]
prp(tree, branch.col=cols, box.col=cols, ...)
}
data(ptitanic)
tree <- rpart(age ~ ., data=ptitanic)
heat.tree(tree, type=4, varlen=0, faclen=0, fallen.leaves=TRUE)

R ML
http://machinelearningmastery.com/machine-learning-in-r-step-by-step/

dendrograms
R Data Viz.
colored dendrogram in R.
# load code of A2R function
source("https://lnkd.in/gkjzrrE")
A2Rplot...
ref: https://rpubs.com/gaston/dendrograms

dendrograms
http://www.sthda.com/english/wiki/beautiful-dendrogram-visualizations-in-r-5-must-known-methods-unsupervised-machine-learning

dendrograms
http://www.sthda.com/english/wiki/beautiful-dendrogram-visualizations-in-r-5-must-known-methods-unsupervised-machine-learning

dendrograms
http://www.sthda.com/english/wiki/beautiful-dendrogram-visualizations-in-r-5-must-known-methods-unsupervised-machine-learning

dendrograms
https://rpubs.com/gaston/dendrograms
http://www.sthda.com/english/wiki/beautiful-dendrogram-visualizations-in-r-5-must-known-methods-unsupervised-machine-learning
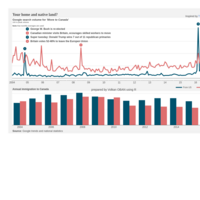
plotly-zoo-time series
library(plotly)
library(zoo)
# Trends Data
trends <- read.csv("https://cdn.rawgit.com/plotly/datasets/master/Move%20to%20Canada.csv", check.names = F, stringsAsFactors = F)
trends.zoo <- zoo(trends[,-1], order.by = as.Date(trends[,1], format = "%d/%m/%Y"))
trends.zoo <- aggregate(trends.zoo, as.yearmon, mean)
trends <- data.frame(Date = index(trends.zoo),
coredata(trends.zoo))
# Immigration Data
immi <- read.csv("https://cdn.rawgit.com/plotly/datasets/master/Canada%20Immigration.csv", stringsAsFactors = F)
labels <- format(as.yearmon(trends$Date), "%Y")
labels <- as.character(sapply(labels, function(x){
unlist(strsplit(x, "20"))[2]
}))
test <- labels[1]
for(i in 2:length(labels)){
if(labels[i] == test) {
labels[i] <- ""
}else{
test <- labels[i]
}
}
labels[1] <- "2004"
hovertext1 <- paste0("Date:<b>", trends$Date, "</b><br>",
"From US:<b>", trends$From.US, "</b><br>")
hovertext2 <- paste0("Date:<b>", trends$Date, "</b><br>",
"From Britain:<b>", trends$From.Britain, "</b><br>")
p <- plot_ly(data = trends, x = ~Date) %>%
# Time series chart
add_lines(y = ~From.US, line = list(color = "#00526d", width = 4),
hoverinfo = "text", text = hovertext1, name = "From US") %>%
add_lines(y = ~From.Britain, line = list(color = "#de6e6e", width = 4),
hoverinfo = "text", text = hovertext2, name = "From Britain") %>%
add_markers(x = c(as.yearmon("2004-11-01"), as.yearmon("2016-03-01")),
y = c(24, 44),
marker = list(size = 15, color = "#00526d"),
showlegend = F) %>%
add_markers(x = c(as.yearmon("2008-07-01"), as.yearmon("2016-07-01")),
y = c(27, 45),
marker = list(size = 15, color = "#de6e6e"),
showlegend = F) %>%
# Markers for legend
add_markers(x = c(as.yearmon("2005-01-01"), as.yearmon("2005-01-01")),
y = c(40, 33.33),
marker = list(size = 15, color = "#00526d"),
showlegend = F) %>%
add_markers(x = c(as.yearmon("2005-01-01"), as.yearmon("2005-01-01")),
y = c(36.67, 30),
marker = list(size = 15, color = "#de6e6e"),
showlegend = F) %>%
add_text(x = c(as.yearmon("2004-11-01"), as.yearmon("2016-03-01")),
y = c(24, 44),
text = c("<b>1</b>", "<b>3</b>"),
textfont = list(color = "white", size = 8),
showlegend = F) %>%
add_text(x = c(as.yearmon("2008-07-01"), as.yearmon("2016-07-01")),
y = c(27, 45),
text = c("<b>2</b>", "<b>4</b>"),
textfont = list(color = "white", size = 8),
showlegend = F) %>%
# Text for legend
add_text(x = c(as.yearmon("2005-01-01"), as.yearmon("2005-01-01"), as.yearmon("2005-01-01"), as.yearmon("2005-01-01")),
y = c(40, 36.67, 33.33, 30),
text = c("<b>1</b>", "<b>2</b>", "<b>3</b>", "<b>4</b>"),
textfont = list(color = "white", size = 8),
showlegend = F) %>%
# Bar chart
add_bars(data = immi, x = ~Year, y = ~USA, yaxis = "y2", xaxis = "x2", showlegend = F,
marker = list(color = "#00526d"), name = "USA") %>%
add_bars(data = immi, x = ~Year, y = ~UK, yaxis = "y2", xaxis = "x2", showlegend = F,
marker = list(color = "#de6e6e"), name = "UK") %>%
layout(legend = list(x = 0.8, y = 0.36, orientation = "h", font = list(size = 10),
bgcolor = "transparent"),
yaxis = list(domain = c(0.4, 0.95), side = "right", title = "", ticklen = 0,
gridwidth = 2),
xaxis = list(showgrid = F, ticklen = 4, nticks = 100,
ticks = "outside",
tickmode = "array",
tickvals = trends$Date,
ticktext = labels,
tickangle = 0,
title = ""),
yaxis2 = list(domain = c(0, 0.3), gridwidth = 2, side = "right"),
xaxis2 = list(anchor = "free", position = 0),
# Annotations
annotations = list(
list(xref = "paper", yref = "paper", xanchor = "left", yanchor = "right",
x = 0, y = 1, showarrow = F,
text = "<b>Your home and native land?</b>",
font = list(size = 18, family = "Balto")),
list(xref = "paper", yref = "paper", xanchor = "left", yanchor = "right",
x = 0, y = 0.95, showarrow = F,
align = "left",
text = "<b>Google search volume for <i>'Move to Canada'</i></b><br><sup>100 is peak volume<br><b>Note</b> that monthly averages are used</sup>",
font = list(size = 13, family = "Arial")),
list(xref = "plot", yref = "plot", xanchor = "left", yanchor = "right",
x = as.yearmon("2005-03-01"), y = 40, showarrow = F,
align = "left",
text = "<b>George W. Bush is re-elected</b>",
font = list(size = 12, family = "Arial"),
bgcolor = "white"),
list(xref = "plot", yref = "plot", xanchor = "left", yanchor = "right",
x = as.yearmon("2005-03-01"), y = 36.67, showarrow = F,
align = "left",
text = "<b>Canadian minister visits Britain, ecourages skilled workers to move</b>",
font = list(size = 12, family = "Arial"),
bgcolor = "white"),
list(xref = "plot", yref = "plot", xanchor = "left", yanchor = "right",
x = as.yearmon("2005-03-01"), y = 33.33, showarrow = F,
align = "left",
text = "<b>Super tuesday: Donald Trump wins 7 out of 11 republican primaries</b>",
font = list(size = 12, family = "Arial"),
bgcolor = "white"),
list(xref = "plot", yref = "plot", xanchor = "left", yanchor = "right",
x = as.yearmon("2005-03-01"), y = 30, showarrow = F,
align = "left",
text = "<b>Britain votes 52-48% to leave the Europen Union</b>",
font = list(size = 12, family = "Arial"),
bgcolor = "white"),
list(xref = "paper", yref = "paper", xanchor = "left", yanchor = "right",
x = 0, y = 0.3, showarrow = F,
align = "left",
text = "<b>Annual immigration to Canada</b>",
font = list(size = 12, family = "Arial")),
list(xref = "paper", yref = "paper", xanchor = "left", yanchor = "right",
x = 0, y = -0.07, showarrow = F,
align = "left",
text = "<b>Source:</b> Google trends and national statistics",
font = list(size = 12, family = "Arial")),
list(xref = "paper", yref = "paper", xanchor = "left", yanchor = "right",
x = 0.85, y = 0.98, showarrow = F,
align = "left",
text = 'Inspired by <a href = "http://www.economist.com/blogs/graphicdetail/2016/07/daily-chart">The economist</a>',
font = list(size = 12, family = "Arial"))),
paper_bgcolor = "#f2f2f2",
margin = list(l = 18, r = 30, t = 18),
width = 1024,height = 600)
print(p)

kNN and plotting
library(MASS)
library(RColorBrewer)
library(class)
mycols <- brewer.pal(8, "Dark2")[c(3,2)]
sink("classification-out.txt")
#########
# Plots showing decision boundaries
s <- sqrt(1/5)
set.seed(30)
makeX <- function(M, n=100, sigma=diag(2)*s) {
z <- sample(1:nrow(M), n, replace=TRUE)
m <- M[z,]
return(t(apply(m,1,function(mu) mvrnorm(1,mu,sigma))))
}
M0 <- mvrnorm(10, c(1,0), diag(2)) # generate 10 means
x0 <- makeX(M0) ## the final values for y0=blue
M1 <- mvrnorm(10, c(0,1), diag(2))
x1 <- makeX(M1)
x <- rbind(x0, x1)
y <- c(rep(0,100), rep(1,100))
cols <- mycols[y+1]
GS <- 75 # put data in a Gs x Gs grid
XLIM <- range(x[,1])
tmpx <- seq(XLIM[1], XLIM[2], len=GS)
YLIM <- range(x[,2])
tmpy <- seq(YLIM[1], YLIM[2], len=GS)
newx <- expand.grid(tmpx, tmpy)
# KNN (1)
yhat <- knn(x, newx, y, k=1)
colshat <- mycols[as.numeric(yhat)]
plot(x, xlab="X1", ylab="X2", xlim=XLIM, ylim=YLIM, type="n")
points(newx, col=colshat, pch=".")
contour(tmpx, tmpy, matrix(as.numeric(yhat),GS,GS), levels=c(1,2), add=TRUE, drawlabels=FALSE)
points(x, col=cols)
title("KNN (1)")

Plot
library(rpart)
library(rpart.plot)
data(ptitanic)
set.seed(123)
tree <- rpart(survived ~ ., data = ptitanic, control = rpart.control(cp = 0.0001))
bestcp <- tree$cptable[which.min(tree$cptable[,"xerror"]),"CP"]
# Step3: Prune the tree using the best cp.
tree.pruned <- prune(tree, cp = bestcp)
conf.matrix <- table(ptitanic$survived, predict(tree.pruned,type="class"))
rownames(conf.matrix) <- paste("Actual", rownames(conf.matrix), sep = ":")
colnames(conf.matrix) <- paste("Pred", colnames(conf.matrix), sep = ":")
print(conf.matrix)
plot(tree.pruned)
text(tree.pruned, cex = 0.8, use.n = TRUE, xpd = TRUE)
prp(tree.pruned, faclen = 0, cex = 0.8, extra = 1)

beeswarm
par(mfrow = c(2,3))
beeswarm(distributions, col = 2:4,
main = 'corral = "none" (default)')
beeswarm(distributions, col = 2:4, corral = "gutter",
main = 'corral = "gutter"')
beeswarm(distributions, col = 2:4, corral = "wrap",
main = 'corral = "wrap"')
beeswarm(distributions, col = 2:4, corral = "random",
main = 'corral = "random"')
beeswarm(distributions, col = 2:4, corral = "omit",
main = 'corral = "omit"')

beeswarm
> distributions <- list(runif = runif(100, min = -3, max = 3),
+ rnorm = rnorm(100),
+ rlnorm = rlnorm(100, sdlog = 0.5))
> beeswarm(distributions, xlab="prepared by VOLKAN OBAN using R-beeswarm", col = 2:4)
beeswarm
Make.Funny.Plot <- function(x){
unique.vals <- length(unique(x))
N <- length(x)
N.val <- min(N/20,unique.vals)
if(unique.vals>N.val){
x <- ave(x,cut(x,N.val),FUN=min)
x <- signif(x,4)
}
# construct the outline of the plot
outline <- as.vector(table(x))
outline <- outline/max(outline)
# determine some correction to make the V shape,
# based on the range
y.corr <- diff(range(x))*0.05
# Get the unique values
yval <- sort(unique(x))
plot(c(-1,1),c(min(yval),max(yval)),
type="n",xaxt="n",xlab="")
for(i in 1:length(yval)){
n <- sum(x==yval[i])
x.plot <- seq(-outline[i],outline[i],length=n)
y.plot <- yval[i]+abs(x.plot)*y.corr
points(x.plot,y.plot,pch=19,cex=0.5)
}
}
x <- rnorm(1000)
Make.Funny.Plot(x)
boxplot(x, add = T, at = 0, col="#0000ff22") # my thanks goes to Greg Snow for the tip on the transparency colour (from 2007): https://stat.ethz.ch/pipermail/r-help/2007-October/142934.html

library(beeswarm)
library(beeswarm)
> data(breast)
> beeswarm(time_survival ~ ER, data = breast,
+ pch = 16, pwcol = 1 + as.numeric(event_survival),
+ xlab = "beeswarm package", ylab = "Follow-up time (months)",
+ labels = c("ER neg", "ER pos"))
> legend("topright", legend = c("Yes", "No"),
+ title = "Censored", pch = 16, col = 1:2

ggbeeswarm -ggplot2 packages
p<-ggplot(mapping=aes(labs, dat)) +
+ geom_quasirandom(method='frowney',alpha=.2) +
+ ggtitle('prepared by Volkan OBAN using R \n smiley') + labs(x='') +
+ theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1))
> p

ggbeeswarm package
p2<-ggplot(mapping=aes(labs, dat)) +
+ geom_quasirandom(method='pseudorandom',alpha=.2) +
+ ggtitle('prepared by Volkan OBAN using R \n pseudorandom') + labs(x='') +
+ theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1))

ggbeeswarm package
ggplot(mpg,aes(class, hwy)) + geom_quasirandom() + ggtitle(" prepared in R by Volkan OBAN \n ggbeeswarm package" ) + theme(plot.title = element_text(size = rel(1), colour = "purple"))
ggbeeswarm package
ggplot2
ggplot(mpg,aes(class, hwy)) + geom_beeswarm(cex=1.1) + ggtitle(" prepared in R by Volkan OBAN \n ggbeeswarm package" ) + theme(plot.title = element_text(size = rel(1), colour = "purple"))

ggplot2 and ggthemes
ggplot(economics_long, aes(date, value01)) +
+ geom_line(aes(linetype = variable)) + ggtitle("prepared by Volkan OBAN using R -- ggplot2and ggthemes packages ") + theme_wsj() + scale_colour_wsj("colors6", "") + theme(plot.title = element_text(size = rel(0.5), colour = "blue"))
ggplot2 and ggthemes
ggplot(economics_long, aes(date, value01)) +
+ geom_line(aes(linetype = variable)) + ggtitle("prepared by Volkan OBAN using R -- ggplot2and ggthemes packages ") + theme_wsj() + scale_colour_wsj("colors6", "")
ggplot2 and ggthemes
ggplot(economics_long, aes(date, value01)) +
+ geom_line(aes(linetype = variable)) + ggtitle("prepared by Volkan OBAN using R -- ggplot2and ggthemes packages ") + theme_solarized(light = FALSE) + scale_colour_solarized("red")

ggplot2 ggthemes pack.
ggplot(economics_long, aes(date, value)) +
+ geom_line() +
+ facet_wrap(~variable, scales = "free_y", nrow = 2, switch = "x") +
+ theme(strip.background = element_blank()) + theme_solarized(light = FALSE) +
+ scale_colour_solarized("red")

cluster package-clustplot
data(iris)
iris.x <- iris[, 1:4]
cl3 <- pam(iris.x, 3)$clustering
op <- par(mfrow= c(2,2))
clusplot(iris.x, cl3, color = TRUE)
U <- par("usr")
## zoom in :
rect(0,-1, 2,1, border = "orange", lwd=2)
clusplot(iris.x, cl3, color = TRUE, xlim = c(0,2), ylim = c(-1,1))
box(col="orange",lwd=2); mtext("sub region", font = 4, cex = 2)
## or zoom out :
clusplot(iris.x, cl3, color = TRUE, xlim = c(-4,4), ylim = c(-4,4))
mtext("`super' region", font = 4, cex = 2)
rect(U[1],U[3], U[2],U[4], lwd=2, lty = 3)
https://stat.ethz.ch/R-manual/R-devel/library/cluster/html/clusplot.default.html

GGally package
ggpairs(iris, upper=list(continuous="density"), lower=list(continuous="smooth"))
Plot
dist <- data.frame(value=rnorm(10000, 1:4), group=1:4)
ggplot(dist, aes(x=value, fill=as.factor(group))) + geom_histogram(alpha=0.5) +geom_vline(data = subset(dist, group=="1"), aes(xintercept=median(value)), color="black",linetype="dashed", size=1) + geom_text(data = subset(dist, group =="1"),aes(x=median(value),y=350,label=round(median(value), digit=3)),hjust=-0.2) +facet_grid(.~group)
ggplot2
> dist <- data.frame(value=rnorm(10000, 1:4), group=1:4)
> ggplot(dist, aes(x=group, y=value, color=group)) + geom_jitter(alpha=0.5)

ggplot2
mg <- ggplot(mtcars, aes(x = mpg, y = wt)) + geom_point()
mg + facet_grid(vs + am ~ gear)
mg + facet_grid(vs + am ~ gear, margins = TRUE)

ggplot2
ggplot(mpg, aes(drv, model)) +
geom_point() +
facet_grid(manufacturer ~ ., scales = "free", space = "free") +
theme(strip.text.y = element_text(angle = 0))

ggplot2 and lattice
qplot(circumference,age, data=Orange, geom=c("line","point"), facets=~Tree)

ggplot2
> library(ggplot2)
> boxplot(circumference~Tree, data=Orange)
> qplot(Tree,circumference, data=Orange, geom=c("boxplot","point"))
plotly
library(plotly)
library(quantmod)
# Download some data
getSymbols(Symbols = c("AAPL", "MSFT"))
ds <- data.frame(Date = index(AAPL), AAPL[,6], MSFT[,6])
p <- plot_ly(ds, x = ~Date) %>%
add_lines(y = ~AAPL.Adjusted, name = "Apple") %>%
add_lines(y = ~MSFT.Adjusted, name = "Microsoft") %>%
layout(
title = "Stock Prices",
xaxis = list(
rangeselector = list(
buttons = list(
list(
count = 3,
label = "3 mo",
step = "month",
stepmode = "backward"),
list(
count = 6,
label = "6 mo",
step = "month",
stepmode = "backward"),
list(
count = 1,
label = "1 yr",
step = "year",
stepmode = "backward"),
list(
count = 1,
label = "YTD",
step = "year",
stepmode = "todate"),
list(step = "all"))),
rangeslider = list(type = "date")),
yaxis = list(title = "Price"))

plotly
p <- plot_ly(
+ plotly::hobbs, r = ~r, t = ~t, color = ~nms, alpha = 0.5, type = "scatter"
+ )
> layout(p, title = "prepared by Volkan OBAN using R-plotly \n Hobbs-Pearson Trials", plot_bgcolor = toRGB("blue")

plotly
> library(plotly)
> p <- plot_ly(
+ plotly::mic, r = ~r, t = ~t, color = ~nms, alpha = 0.5, type = "scatter"
+ )
> layout(p, title = "prepared by Volkan OBAN using R-plotly \n Mic Patterns", orientation = -90)

ggpairs
pm = ggpairs(data=tips,
+ columns=1:3,
+ upper = list(continuous = "density"),
+ lower = list(combo = "facetdensity"),
+ title="tips data",
+ colour = "sex")
pm

ggord package
https://github.com/fawda123/ggord
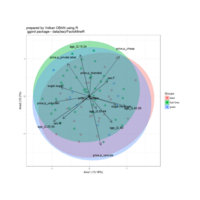
ggord package
https://github.com/fawda123/ggord

ggord package
library(ggord)
library(FactoMineR)
> data("tea")
> tea <- tea[, c('Tea', 'sugar', 'price', 'age_Q', 'sex')]
>
> ord <- MCA(tea[, -1], graph = FALSE)
>
> ggord(ord, tea$Tea)

dotplot
> m3a <- glmer(remission ~ Age + LengthofStay + FamilyHx + IL6 + CRP +
+ CancerStage + Experience + (1 | DID) + (1 | HID),
+ data = hdp, family = binomial, nAGQ=1)
Warning message:
In checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.400012 (tol = 0.001, component 1)
> dotplot(ranef(m3a, which = "DID", postVar = TRUE), scales = list(y = list(alternating = 0)))

Plot
ref. and data:
http://www.ats.ucla.edu/stat/r/dae/melogit.htm

GGally
ref: http://www.ats.ucla.edu/stat/r/dae/melogit.htm

gridExtra-
p2 <- ggplot(mtcars, aes(mpg, wt, colour = factor(cyl))) +
geom_point() + facet_wrap( ~ cyl, ncol=2, scales = "free") +
guides(colour="none") +
theme()
grid.arrange(tableGrob(mtcars[1:4, 1:4]), p2,
ncol=2, widths=c(1.5, 1), clip=FALSE)

Plot
>
> moxbuller = function(n) {
+ u = runif(n)
+ v = runif(n)
+ x = cos(2*pi*u)*sqrt(-2*log(v))
+ y = sin(2*pi*v)*sqrt(-2*log(u))
+ r = list(x=x, y=y)
+ return(r)
+ }
> r = moxbuller(50000)
> par(bg="white")
> par(mar=c(0,0,0,0))
> plot(r$x,r$y, pch=".", col="red", main=" \n \n \n prepared in R by VOLKAN OBAN", cex=1.2)

lattice
library(lattice)
library(psych)
d <- colMeans(msq[,c(2,7,34,36,42,43,46,55,68)],na.rm = T)*10
barchart(sort(d), xlab="", ylab="", col = "grey", origin=1,
border = "transparent", box.ratio=0.5,
panel = function(x,y,...) {
panel.barchart(x,y,...)
panel.abline(v=seq(1,6,1), col="white", lwd=3)},
par.settings = list(axis.line = list(col = "transparent")))
ltext(current.panel.limits()$xlim[2]-50, adj=1,
current.panel.limits()$ylim[1]-100,
"Average scores\non negative emotion traits\nfrom 3896 participants\n(Watson et al., 1988)")
ggplot2
#--Load previously saved data:
path <- "http://www.sr.bham.ac.uk/~ajrs/R/datasets"
a <- load(url(paste(path,"middayweather.RData",sep="/")))
close(url(paste(path,"middayweather.RData",sep="/")))
#--Load extra library:
## if not already installed, then run:
# install.packages("ggplot2")
require(ggplot2)
#--Calculate month from date & create factor:
middayweather$month <- with(middayweather, factor(as.POSIXlt(Date)$mon, label=month.abb))
#--Calculate year from date:
middayweather$year <- with(middayweather, 1900 + as.POSIXlt(Date)$year)
#--Only use complete years:
middayweather <- subset(middayweather, year %in% 2007:2009)
#-----Plot data as boxplot summary for each month:
theme_set(theme_gray(base_size = 11))
p <- ggplot(data=middayweather, aes(month, T.out)) +
geom_boxplot() +
facet_wrap( ~ year, nrow=1) +
scale_x_discrete(breaks=month.abb[c(1, 4, 7, 10)]) +
xlab("") +
ylab(as.expression(expression( paste("Temperature (", degree,"C)") )))

coplot
> coplot(ll.dm, data = quakes, number = c(3, 7),
overlap = c(-.5, .1),xlab = "long", bar.bg = c(fac = "blue"))

coplot lattice
coplot(breaks ~ Index | wool * tension, data = warpbreaks,
+ col = "red", bg = "black", pch = 21,xlab = "Index",
+ bar.bg = c(fac = "purple"))

lattice
> par(mfrow=c(1,3))
> mysplits = split(mtcars,mtcars$cyl)
> maxmpg = max(mtcars$mpg)
> for (ii in 1:length(mysplits)) {
+ tmpdf <- mysplits[[ii]]
+ auto <- tmpdf[tmpdf$am == 0,]
+ man <- tmpdf[tmpdf$am == 1,]
+ plot(tmpdf$wt , tmpdf$mpg,type="n",
+ main=paste(names(mysplits[ii])," Cylinders"),
+ ylim=c(0,maxmpg), xlab="wt",ylab="MPG")
+ points(auto$wt,auto$mpg,col="blue",pch=19)
+ points(man$wt,man$mpg,col="red",pch=19)
+ grid()
+ legend("topright", inset=0.05, c("manual","auto"),
pch = 19, col=c("red","blue"))
}
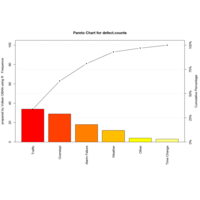
pareto chart
> defect.counts <- c(12,29,18,3,34,4)
> names(defect.counts) <- c("Weather","Overslept", "Alarm Failure",
+ "Time Change","Traffic","Other")
> df.defects <- data.frame(defect.counts)
>
> df.defects
library(qcc)
pareto.chart(defect.counts)

business Analytics graph Example
ref:http://analyzecore.com/2015/04/01/cohort-analysis-and-lifecycle-grids-mixed-segmentation-with-r/

business Analytics graph Example
library(dplyr)
library(reshape2)
library(ggplot2)
library(scales)
library(gridExtra)
# creating data sample
set.seed(10)
cohorts <- data.frame(cohort = paste('cohort', formatC(c(1:36), width=2, format='d', flag='0'), sep = '_'),
Y_00 = sample(c(1300:1500), 36, replace = TRUE),
Y_01 = c(sample(c(800:1000), 36, replace = TRUE)),
Y_02 = c(sample(c(600:800), 24, replace = TRUE), rep(NA, 12)),
Y_03 = c(sample(c(400:500), 12, replace = TRUE), rep(NA, 24)))
# simulating seasonality (Black Friday)
cohorts[c(11, 23, 35), 2] <- as.integer(cohorts[c(11, 23, 35), 2] * 1.25)
cohorts[c(11, 23, 35), 3] <- as.integer(cohorts[c(11, 23, 35), 3] * 1.10)
cohorts[c(11, 23, 35), 4] <- as.integer(cohorts[c(11, 23, 35), 4] * 1.07)
# calculating retention rate and preparing data for plotting
df_plot <- melt(cohorts, id.vars = 'cohort', value.name = 'number', variable.name = 'year_of_LT')
df_plot <- df_plot %>%
group_by(cohort) %>%
arrange(year_of_LT) %>%
mutate(number_prev_year = lag(number),
number_Y_00 = number[which(year_of_LT == 'Y_00')]) %>%
ungroup() %>%
mutate(ret_rate_prev_year = number / number_prev_year,
ret_rate = number / number_Y_00,
year_cohort = paste(year_of_LT, cohort, sep = '-'))
##### The first way for plotting cycle plot via scaling
# calculating the coefficient for scaling 2nd axis
k <- max(df_plot$number_prev_year[df_plot$year_of_LT == 'Y_01'] * 1.15) / min(df_plot$ret_rate[df_plot$year_of_LT == 'Y_01'])
# retention rate cycle plot
ggplot(na.omit(df_plot), aes(x = year_cohort, y = ret_rate, group = year_of_LT, color = year_of_LT)) +
theme_bw() +
geom_point(size = 4) +
geom_text(aes(label = percent(round(ret_rate, 2))),
size = 4, hjust = 0.4, vjust = -0.6, fontface = "plain") +
# smooth method can be changed (e.g. for "lm")
geom_smooth(size = 2.5, method = 'loess', color = 'darkred', aes(fill = year_of_LT)) +
geom_bar(aes(y = number_prev_year / k, fill = year_of_LT), alpha = 0.2, stat = 'identity') +
geom_bar(aes(y = number / k, fill = year_of_LT), alpha = 0.6, stat = 'identity') +
geom_text(aes(y = 0, label = cohort), color = 'white', angle = 90, size = 4, hjust = -0.05, vjust = 0.4) +
geom_text(aes(y = number_prev_year / k, label = number_prev_year),
angle = 90, size = 4, hjust = -0.1, vjust = 0.4) +
geom_text(aes(y = number / k, label = number),
angle = 90, size = 4, hjust = -0.1, vjust = 0.4) +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_text(size=18, face="bold"),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_text(size=16),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(x = 'Year of Lifetime by Cohorts', y = 'Number of Customers / Retention Rate') +
ggtitle("Customer Retention Rate - Cycle plot")
##### The second way for plotting cycle plot via multi-plotting
# plot #1 - Retention rate
p1 <- ggplot(na.omit(df_plot), aes(x = year_cohort, y = ret_rate, group = year_of_LT, color = year_of_LT)) +
theme_bw() +
geom_point(size = 4) +
geom_text(aes(label = percent(round(ret_rate, 2))),
size = 4, hjust = 0.4, vjust = -0.6, fontface = "plain") +
geom_smooth(size = 2.5, method = 'loess', color = 'darkred', aes(fill = year_of_LT)) +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_blank(),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(y = 'Retention Rate') +
ggtitle("Customer Retention Rate - Cycle plot")
# plot #2 - number of customers
p2 <- ggplot(na.omit(df_plot), aes(x = year_cohort, group = year_of_LT, color = year_of_LT)) +
theme_bw() +
geom_bar(aes(y = number_prev_year, fill = year_of_LT), alpha = 0.2, stat = 'identity') +
geom_bar(aes(y = number, fill = year_of_LT), alpha = 0.6, stat = 'identity') +
geom_text(aes(y = number_prev_year, label = number_prev_year),
angle = 90, size = 4, hjust = -0.1, vjust = 0.4) +
geom_text(aes(y = number, label = number),
angle = 90, size = 4, hjust = -0.1, vjust = 0.4) +
geom_text(aes(y = 0, label = cohort), color = 'white', angle = 90, size = 4, hjust = -0.05, vjust = 0.4) +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_text(size=18, face="bold"),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
scale_y_continuous(limits = c(0, max(df_plot$number_Y_00 * 1.1))) +
labs(x = 'Year of Lifetime by Cohorts', y = 'Number of Customers')
# multiplot
grid.arrange(p1, p2, ncol = 1)
# retention rate bubble chart
ggplot(na.omit(df_plot), aes(x = cohort, y = ret_rate, group = cohort, color = year_of_LT)) +
theme_bw() +
scale_size(range = c(15, 40)) +
geom_line(size = 2, alpha = 0.3) +
geom_point(aes(size = number_prev_year), alpha = 0.3) +
geom_point(aes(size = number), alpha = 0.8) +
geom_smooth(linetype = 2, size = 2, method = 'loess', aes(group = year_of_LT, fill = year_of_LT), alpha = 0.2) +
geom_text(aes(label = paste0(number, '/', number_prev_year, '\n', percent(round(ret_rate, 2)))),
color = 'white', size = 3, hjust = 0.5, vjust = 0.5, fontface = "plain") +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_text(size=18, face="bold"),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_text(size=16),
axis.text.x = element_text(size=10, angle=90, hjust=.5, vjust=.5, face="plain"),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(x = 'Cohorts', y = 'Retention Rate by Year of Lifetime') +
ggtitle("Customer Retention Rate - Bubble chart")
# retention rate falling drops chart
ggplot(df_plot, aes(x = cohort, y = ret_rate, group = cohort, color = year_of_LT)) +
theme_bw() +
scale_size(range = c(15, 40)) +
scale_y_continuous(limits = c(0, 1)) +
geom_line(size = 2, alpha = 0.3) +
geom_point(aes(size = number), alpha = 0.8) +
geom_text(aes(label = paste0(number, '\n', percent(round(ret_rate, 2)))),
color = 'white', size = 3, hjust = 0.5, vjust = 0.5, fontface = "plain") +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_text(size=18, face="bold"),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_text(size=16),
axis.text.x = element_text(size=10, angle=90, hjust=.5, vjust=.5, face="plain"),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(x = 'Cohorts', y = 'Retention Rate by Year of Lifetime') +
ggtitle("Customer Retention Rate - Falling Drops chart")
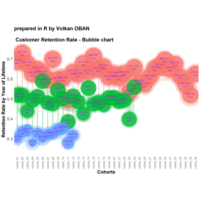
Bus.analytics graphs
library(dplyr)
library(reshape2)
library(ggplot2)
library(scales)
library(gridExtra)
# creating data sample
set.seed(10)
cohorts <- data.frame(cohort = paste('cohort', formatC(c(1:36), width=2, format='d', flag='0'), sep = '_'),
Y_00 = sample(c(1300:1500), 36, replace = TRUE),
Y_01 = c(sample(c(800:1000), 36, replace = TRUE)),
Y_02 = c(sample(c(600:800), 24, replace = TRUE), rep(NA, 12)),
Y_03 = c(sample(c(400:500), 12, replace = TRUE), rep(NA, 24)))
# simulating seasonality (Black Friday)
cohorts[c(11, 23, 35), 2] <- as.integer(cohorts[c(11, 23, 35), 2] * 1.25)
cohorts[c(11, 23, 35), 3] <- as.integer(cohorts[c(11, 23, 35), 3] * 1.10)
cohorts[c(11, 23, 35), 4] <- as.integer(cohorts[c(11, 23, 35), 4] * 1.07)
# calculating retention rate and preparing data for plotting
df_plot <- melt(cohorts, id.vars = 'cohort', value.name = 'number', variable.name = 'year_of_LT')
df_plot <- df_plot %>%
group_by(cohort) %>%
arrange(year_of_LT) %>%
mutate(number_prev_year = lag(number),
number_Y_00 = number[which(year_of_LT == 'Y_00')]) %>%
ungroup() %>%
mutate(ret_rate_prev_year = number / number_prev_year,
ret_rate = number / number_Y_00,
year_cohort = paste(year_of_LT, cohort, sep = '-'))
##### The first way for plotting cycle plot via scaling
# calculating the coefficient for scaling 2nd axis
k <- max(df_plot$number_prev_year[df_plot$year_of_LT == 'Y_01'] * 1.15) / min(df_plot$ret_rate[df_plot$year_of_LT == 'Y_01'])
# retention rate cycle plot
ggplot(na.omit(df_plot), aes(x = year_cohort, y = ret_rate, group = year_of_LT, color = year_of_LT)) +
theme_bw() +
geom_point(size = 4) +
geom_text(aes(label = percent(round(ret_rate, 2))),
size = 4, hjust = 0.4, vjust = -0.6, fontface = "plain") +
# smooth method can be changed (e.g. for "lm")
geom_smooth(size = 2.5, method = 'loess', color = 'darkred', aes(fill = year_of_LT)) +
geom_bar(aes(y = number_prev_year / k, fill = year_of_LT), alpha = 0.2, stat = 'identity') +
geom_bar(aes(y = number / k, fill = year_of_LT), alpha = 0.6, stat = 'identity') +
geom_text(aes(y = 0, label = cohort), color = 'white', angle = 90, size = 4, hjust = -0.05, vjust = 0.4) +
geom_text(aes(y = number_prev_year / k, label = number_prev_year),
angle = 90, size = 4, hjust = -0.1, vjust = 0.4) +
geom_text(aes(y = number / k, label = number),
angle = 90, size = 4, hjust = -0.1, vjust = 0.4) +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_text(size=18, face="bold"),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_text(size=16),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(x = 'Year of Lifetime by Cohorts', y = 'Number of Customers / Retention Rate') +
ggtitle("Customer Retention Rate - Cycle plot")
##### The second way for plotting cycle plot via multi-plotting
# plot #1 - Retention rate
p1 <- ggplot(na.omit(df_plot), aes(x = year_cohort, y = ret_rate, group = year_of_LT, color = year_of_LT)) +
theme_bw() +
geom_point(size = 4) +
geom_text(aes(label = percent(round(ret_rate, 2))),
size = 4, hjust = 0.4, vjust = -0.6, fontface = "plain") +
geom_smooth(size = 2.5, method = 'loess', color = 'darkred', aes(fill = year_of_LT)) +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_blank(),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(y = 'Retention Rate') +
ggtitle("Customer Retention Rate - Cycle plot")
# plot #2 - number of customers
p2 <- ggplot(na.omit(df_plot), aes(x = year_cohort, group = year_of_LT, color = year_of_LT)) +
theme_bw() +
geom_bar(aes(y = number_prev_year, fill = year_of_LT), alpha = 0.2, stat = 'identity') +
geom_bar(aes(y = number, fill = year_of_LT), alpha = 0.6, stat = 'identity') +
geom_text(aes(y = number_prev_year, label = number_prev_year),
angle = 90, size = 4, hjust = -0.1, vjust = 0.4) +
geom_text(aes(y = number, label = number),
angle = 90, size = 4, hjust = -0.1, vjust = 0.4) +
geom_text(aes(y = 0, label = cohort), color = 'white', angle = 90, size = 4, hjust = -0.05, vjust = 0.4) +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_text(size=18, face="bold"),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
scale_y_continuous(limits = c(0, max(df_plot$number_Y_00 * 1.1))) +
labs(x = 'Year of Lifetime by Cohorts', y = 'Number of Customers')
# multiplot
grid.arrange(p1, p2, ncol = 1)
# retention rate bubble chart
ggplot(na.omit(df_plot), aes(x = cohort, y = ret_rate, group = cohort, color = year_of_LT)) +
theme_bw() +
scale_size(range = c(15, 40)) +
geom_line(size = 2, alpha = 0.3) +
geom_point(aes(size = number_prev_year), alpha = 0.3) +
geom_point(aes(size = number), alpha = 0.8) +
geom_smooth(linetype = 2, size = 2, method = 'loess', aes(group = year_of_LT, fill = year_of_LT), alpha = 0.2) +
geom_text(aes(label = paste0(number, '/', number_prev_year, '\n', percent(round(ret_rate, 2)))),
color = 'white', size = 3, hjust = 0.5, vjust = 0.5, fontface = "plain") +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_text(size=18, face="bold"),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_text(size=16),
axis.text.x = element_text(size=10, angle=90, hjust=.5, vjust=.5, face="plain"),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(x = 'Cohorts', y = 'Retention Rate by Year of Lifetime') +
ggtitle("Customer Retention Rate - Bubble chart")
# retention rate falling drops chart
ggplot(df_plot, aes(x = cohort, y = ret_rate, group = cohort, color = year_of_LT)) +
theme_bw() +
scale_size(range = c(15, 40)) +
scale_y_continuous(limits = c(0, 1)) +
geom_line(size = 2, alpha = 0.3) +
geom_point(aes(size = number), alpha = 0.8) +
geom_text(aes(label = paste0(number, '\n', percent(round(ret_rate, 2)))),
color = 'white', size = 3, hjust = 0.5, vjust = 0.5, fontface = "plain") +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_text(size=18, face="bold"),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_text(size=16),
axis.text.x = element_text(size=10, angle=90, hjust=.5, vjust=.5, face="plain"),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(x = 'Cohorts', y = 'Retention Rate by Year of Lifetime') +
ggtitle("Customer Retention Rate - Falling Drops chart")

Bus.analytics graphs
library(dplyr)
library(reshape2)
library(ggplot2)
library(scales)
library(gridExtra)
# creating data sample
set.seed(10)
cohorts <- data.frame(cohort = paste('cohort', formatC(c(1:36), width=2, format='d', flag='0'), sep = '_'),
Y_00 = sample(c(1300:1500), 36, replace = TRUE),
Y_01 = c(sample(c(800:1000), 36, replace = TRUE)),
Y_02 = c(sample(c(600:800), 24, replace = TRUE), rep(NA, 12)),
Y_03 = c(sample(c(400:500), 12, replace = TRUE), rep(NA, 24)))
# simulating seasonality (Black Friday)
cohorts[c(11, 23, 35), 2] <- as.integer(cohorts[c(11, 23, 35), 2] * 1.25)
cohorts[c(11, 23, 35), 3] <- as.integer(cohorts[c(11, 23, 35), 3] * 1.10)
cohorts[c(11, 23, 35), 4] <- as.integer(cohorts[c(11, 23, 35), 4] * 1.07)
# calculating retention rate and preparing data for plotting
df_plot <- melt(cohorts, id.vars = 'cohort', value.name = 'number', variable.name = 'year_of_LT')
df_plot <- df_plot %>%
group_by(cohort) %>%
arrange(year_of_LT) %>%
mutate(number_prev_year = lag(number),
number_Y_00 = number[which(year_of_LT == 'Y_00')]) %>%
ungroup() %>%
mutate(ret_rate_prev_year = number / number_prev_year,
ret_rate = number / number_Y_00,
year_cohort = paste(year_of_LT, cohort, sep = '-'))
##### The first way for plotting cycle plot via scaling
# calculating the coefficient for scaling 2nd axis
k <- max(df_plot$number_prev_year[df_plot$year_of_LT == 'Y_01'] * 1.15) / min(df_plot$ret_rate[df_plot$year_of_LT == 'Y_01'])
# retention rate cycle plot
ggplot(na.omit(df_plot), aes(x = year_cohort, y = ret_rate, group = year_of_LT, color = year_of_LT)) +
theme_bw() +
geom_point(size = 4) +
geom_text(aes(label = percent(round(ret_rate, 2))),
size = 4, hjust = 0.4, vjust = -0.6, fontface = "plain") +
# smooth method can be changed (e.g. for "lm")
geom_smooth(size = 2.5, method = 'loess', color = 'darkred', aes(fill = year_of_LT)) +
geom_bar(aes(y = number_prev_year / k, fill = year_of_LT), alpha = 0.2, stat = 'identity') +
geom_bar(aes(y = number / k, fill = year_of_LT), alpha = 0.6, stat = 'identity') +
geom_text(aes(y = 0, label = cohort), color = 'white', angle = 90, size = 4, hjust = -0.05, vjust = 0.4) +
geom_text(aes(y = number_prev_year / k, label = number_prev_year),
angle = 90, size = 4, hjust = -0.1, vjust = 0.4) +
geom_text(aes(y = number / k, label = number),
angle = 90, size = 4, hjust = -0.1, vjust = 0.4) +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_text(size=18, face="bold"),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_text(size=16),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(x = 'Year of Lifetime by Cohorts', y = 'Number of Customers / Retention Rate') +
ggtitle("Customer Retention Rate - Cycle plot")
##### The second way for plotting cycle plot via multi-plotting
# plot #1 - Retention rate
p1 <- ggplot(na.omit(df_plot), aes(x = year_cohort, y = ret_rate, group = year_of_LT, color = year_of_LT)) +
theme_bw() +
geom_point(size = 4) +
geom_text(aes(label = percent(round(ret_rate, 2))),
size = 4, hjust = 0.4, vjust = -0.6, fontface = "plain") +
geom_smooth(size = 2.5, method = 'loess', color = 'darkred', aes(fill = year_of_LT)) +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_blank(),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(y = 'Retention Rate') +
ggtitle("Customer Retention Rate - Cycle plot")
# plot #2 - number of customers
p2 <- ggplot(na.omit(df_plot), aes(x = year_cohort, group = year_of_LT, color = year_of_LT)) +
theme_bw() +
geom_bar(aes(y = number_prev_year, fill = year_of_LT), alpha = 0.2, stat = 'identity') +
geom_bar(aes(y = number, fill = year_of_LT), alpha = 0.6, stat = 'identity') +
geom_text(aes(y = number_prev_year, label = number_prev_year),
angle = 90, size = 4, hjust = -0.1, vjust = 0.4) +
geom_text(aes(y = number, label = number),
angle = 90, size = 4, hjust = -0.1, vjust = 0.4) +
geom_text(aes(y = 0, label = cohort), color = 'white', angle = 90, size = 4, hjust = -0.05, vjust = 0.4) +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_text(size=18, face="bold"),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
scale_y_continuous(limits = c(0, max(df_plot$number_Y_00 * 1.1))) +
labs(x = 'Year of Lifetime by Cohorts', y = 'Number of Customers')
# multiplot
grid.arrange(p1, p2, ncol = 1)
# retention rate bubble chart
ggplot(na.omit(df_plot), aes(x = cohort, y = ret_rate, group = cohort, color = year_of_LT)) +
theme_bw() +
scale_size(range = c(15, 40)) +
geom_line(size = 2, alpha = 0.3) +
geom_point(aes(size = number_prev_year), alpha = 0.3) +
geom_point(aes(size = number), alpha = 0.8) +
geom_smooth(linetype = 2, size = 2, method = 'loess', aes(group = year_of_LT, fill = year_of_LT), alpha = 0.2) +
geom_text(aes(label = paste0(number, '/', number_prev_year, '\n', percent(round(ret_rate, 2)))),
color = 'white', size = 3, hjust = 0.5, vjust = 0.5, fontface = "plain") +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_text(size=18, face="bold"),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_text(size=16),
axis.text.x = element_text(size=10, angle=90, hjust=.5, vjust=.5, face="plain"),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(x = 'Cohorts', y = 'Retention Rate by Year of Lifetime') +
ggtitle("Customer Retention Rate - Bubble chart")
# retention rate falling drops chart
ggplot(df_plot, aes(x = cohort, y = ret_rate, group = cohort, color = year_of_LT)) +
theme_bw() +
scale_size(range = c(15, 40)) +
scale_y_continuous(limits = c(0, 1)) +
geom_line(size = 2, alpha = 0.3) +
geom_point(aes(size = number), alpha = 0.8) +
geom_text(aes(label = paste0(number, '\n', percent(round(ret_rate, 2)))),
color = 'white', size = 3, hjust = 0.5, vjust = 0.5, fontface = "plain") +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_text(size=18, face="bold"),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_text(size=16),
axis.text.x = element_text(size=10, angle=90, hjust=.5, vjust=.5, face="plain"),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(x = 'Cohorts', y = 'Retention Rate by Year of Lifetime') +
ggtitle("Customer Retention Rate - Falling Drops chart")
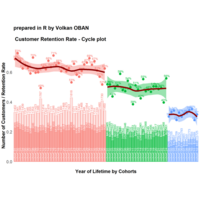
Bus.analytics graphs
library(dplyr)
library(reshape2)
library(ggplot2)
library(scales)
library(gridExtra)
# creating data sample
set.seed(10)
cohorts <- data.frame(cohort = paste('cohort', formatC(c(1:36), width=2, format='d', flag='0'), sep = '_'),
Y_00 = sample(c(1300:1500), 36, replace = TRUE),
Y_01 = c(sample(c(800:1000), 36, replace = TRUE)),
Y_02 = c(sample(c(600:800), 24, replace = TRUE), rep(NA, 12)),
Y_03 = c(sample(c(400:500), 12, replace = TRUE), rep(NA, 24)))
# simulating seasonality (Black Friday)
cohorts[c(11, 23, 35), 2] <- as.integer(cohorts[c(11, 23, 35), 2] * 1.25)
cohorts[c(11, 23, 35), 3] <- as.integer(cohorts[c(11, 23, 35), 3] * 1.10)
cohorts[c(11, 23, 35), 4] <- as.integer(cohorts[c(11, 23, 35), 4] * 1.07)
# calculating retention rate and preparing data for plotting
df_plot <- melt(cohorts, id.vars = 'cohort', value.name = 'number', variable.name = 'year_of_LT')
df_plot <- df_plot %>%
group_by(cohort) %>%
arrange(year_of_LT) %>%
mutate(number_prev_year = lag(number),
number_Y_00 = number[which(year_of_LT == 'Y_00')]) %>%
ungroup() %>%
mutate(ret_rate_prev_year = number / number_prev_year,
ret_rate = number / number_Y_00,
year_cohort = paste(year_of_LT, cohort, sep = '-'))
##### The first way for plotting cycle plot via scaling
# calculating the coefficient for scaling 2nd axis
k <- max(df_plot$number_prev_year[df_plot$year_of_LT == 'Y_01'] * 1.15) / min(df_plot$ret_rate[df_plot$year_of_LT == 'Y_01'])
# retention rate cycle plot
ggplot(na.omit(df_plot), aes(x = year_cohort, y = ret_rate, group = year_of_LT, color = year_of_LT)) +
theme_bw() +
geom_point(size = 4) +
geom_text(aes(label = percent(round(ret_rate, 2))),
size = 4, hjust = 0.4, vjust = -0.6, fontface = "plain") +
# smooth method can be changed (e.g. for "lm")
geom_smooth(size = 2.5, method = 'loess', color = 'darkred', aes(fill = year_of_LT)) +
geom_bar(aes(y = number_prev_year / k, fill = year_of_LT), alpha = 0.2, stat = 'identity') +
geom_bar(aes(y = number / k, fill = year_of_LT), alpha = 0.6, stat = 'identity') +
geom_text(aes(y = 0, label = cohort), color = 'white', angle = 90, size = 4, hjust = -0.05, vjust = 0.4) +
geom_text(aes(y = number_prev_year / k, label = number_prev_year),
angle = 90, size = 4, hjust = -0.1, vjust = 0.4) +
geom_text(aes(y = number / k, label = number),
angle = 90, size = 4, hjust = -0.1, vjust = 0.4) +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_text(size=18, face="bold"),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_text(size=16),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(x = 'Year of Lifetime by Cohorts', y = 'Number of Customers / Retention Rate') +
ggtitle("Customer Retention Rate - Cycle plot")
##### The second way for plotting cycle plot via multi-plotting
# plot #1 - Retention rate
p1 <- ggplot(na.omit(df_plot), aes(x = year_cohort, y = ret_rate, group = year_of_LT, color = year_of_LT)) +
theme_bw() +
geom_point(size = 4) +
geom_text(aes(label = percent(round(ret_rate, 2))),
size = 4, hjust = 0.4, vjust = -0.6, fontface = "plain") +
geom_smooth(size = 2.5, method = 'loess', color = 'darkred', aes(fill = year_of_LT)) +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_blank(),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(y = 'Retention Rate') +
ggtitle("Customer Retention Rate - Cycle plot")
# plot #2 - number of customers
p2 <- ggplot(na.omit(df_plot), aes(x = year_cohort, group = year_of_LT, color = year_of_LT)) +
theme_bw() +
geom_bar(aes(y = number_prev_year, fill = year_of_LT), alpha = 0.2, stat = 'identity') +
geom_bar(aes(y = number, fill = year_of_LT), alpha = 0.6, stat = 'identity') +
geom_text(aes(y = number_prev_year, label = number_prev_year),
angle = 90, size = 4, hjust = -0.1, vjust = 0.4) +
geom_text(aes(y = number, label = number),
angle = 90, size = 4, hjust = -0.1, vjust = 0.4) +
geom_text(aes(y = 0, label = cohort), color = 'white', angle = 90, size = 4, hjust = -0.05, vjust = 0.4) +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_text(size=18, face="bold"),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
scale_y_continuous(limits = c(0, max(df_plot$number_Y_00 * 1.1))) +
labs(x = 'Year of Lifetime by Cohorts', y = 'Number of Customers')
# multiplot
grid.arrange(p1, p2, ncol = 1)
# retention rate bubble chart
ggplot(na.omit(df_plot), aes(x = cohort, y = ret_rate, group = cohort, color = year_of_LT)) +
theme_bw() +
scale_size(range = c(15, 40)) +
geom_line(size = 2, alpha = 0.3) +
geom_point(aes(size = number_prev_year), alpha = 0.3) +
geom_point(aes(size = number), alpha = 0.8) +
geom_smooth(linetype = 2, size = 2, method = 'loess', aes(group = year_of_LT, fill = year_of_LT), alpha = 0.2) +
geom_text(aes(label = paste0(number, '/', number_prev_year, '\n', percent(round(ret_rate, 2)))),
color = 'white', size = 3, hjust = 0.5, vjust = 0.5, fontface = "plain") +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_text(size=18, face="bold"),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_text(size=16),
axis.text.x = element_text(size=10, angle=90, hjust=.5, vjust=.5, face="plain"),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(x = 'Cohorts', y = 'Retention Rate by Year of Lifetime') +
ggtitle("Customer Retention Rate - Bubble chart")
# retention rate falling drops chart
ggplot(df_plot, aes(x = cohort, y = ret_rate, group = cohort, color = year_of_LT)) +
theme_bw() +
scale_size(range = c(15, 40)) +
scale_y_continuous(limits = c(0, 1)) +
geom_line(size = 2, alpha = 0.3) +
geom_point(aes(size = number), alpha = 0.8) +
geom_text(aes(label = paste0(number, '\n', percent(round(ret_rate, 2)))),
color = 'white', size = 3, hjust = 0.5, vjust = 0.5, fontface = "plain") +
theme(legend.position='none',
plot.title = element_text(size=20, face="bold", vjust=2),
axis.title.x = element_text(size=18, face="bold"),
axis.title.y = element_text(size=18, face="bold"),
axis.text = element_text(size=16),
axis.text.x = element_text(size=10, angle=90, hjust=.5, vjust=.5, face="plain"),
axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(x = 'Cohorts', y = 'Retention Rate by Year of Lifetime') +
ggtitle("Customer Retention Rate - Falling Drops chart")

cohort analysis in R
cohort.sum <- data.frame(cohort=c('Cohort01', 'Cohort02', 'Cohort03', 'Cohort04', 'Cohort05', 'Cohort06', 'Cohort07', 'Cohort08', 'Cohort09', 'Cohort10', 'Cohort11', 'Cohort12'),
M1=c(270000,0,0,0,0,0,0,0,0,0,0,0),
M2=c(85000,275000,0,0,0,0,0,0,0,0,0,0),
M3=c(72000,63000,277000,0,0,0,0,0,0,0,0,0),
M4=c(52000,42000,76000,361000,0,0,0,0,0,0,0,0),
M5=c(50000,45000,60000,80000,288000,0,0,0,0,0,0,0),
M6=c(51000,52000,55000,51000,58000,253000,0,0,0,0,0,0),
M7=c(51000,69000,48000,45000,42000,54000,272000,0,0,0,0,0),
M8=c(46000,85000,77000,41000,38000,37000,74000,352000,0,0,0,0),
M9=c(38000,42000,72000,41000,31000,30000,49000,107000,285000,0,0,0),
M10=c(39000,38000,45000,33000,34000,34000,46000,83000,69000,279000,0,0),
M11=c(38000,42000,31000,32000,26000,28000,43000,82000,51000,87000,282000,0),
M12=c(35000,35000,38000,45000,35000,32000,48000,44000,47000,52000,92000,500000))
ibrary(ggplot2)
library(reshape2)
#we need to melt data
cohort.chart <- melt(cohort.sum, id.vars = "cohort")
colnames(cohort.chart) <- c('cohort', 'month', 'revenue')
#define palette
blues <- colorRampPalette(c('red', 'black'))
#plot data
p <- ggplot(cohort.chart, aes(x=month, y=revenue, group=cohort))
p + geom_area(aes(fill = cohort)) +
scale_fill_manual(values = blues(nrow(cohort.sum))) +
ggtitle('Total revenue by Cohort')
lattice-ggplot2-rbokeh
bp <- figure( height = 400, width = 700 )
data(barley,package="lattice")
# no facet to get started
bp %>%
ly_points( yield, variety, barley, color = year, hover = list( variety, yield ) )
# now show facet
lapply(
levels( barley$site )
,function(s){
figure( height = 200, width = 700, title = s ) %>%
ly_points(
yield, variety
# for fun do without dplyr
, data = subset(barley, site == s)
, color = year
, hover = list( variety, yield )
, size = 6
)
}
) %>%
grid_plot( nrow = length(.), ncol = 1, same_axes = T )

rbokeh
library(rbokeh)
library(ggplot2)
data(diamonds)
bp <- figure( height = 400, width = 700 )
bp %>% ly_points(carat,price,diamonds,color=cut,size=5)
# add hover
bp %>%
ly_points(carat,price,diamonds,color=cut,size=1,hover=list(cut,clarity,color))
# make it a hexbin
bp %>%
ly_hexbin( carat, price, diamonds )
# make it a hexbin with facet by color
lapply(
levels(unique(diamonds$color))
,function(c){
figure( height = 300, width = 300, title = paste0("Color: ",c) ) %>%
ly_hexbin( carat, log(price), diamonds[which(diamonds$color==c),] )
}
) %>%
grid_plot( nrow = 3, ncol = 3, same_axes = T )
# histogram on diamonds
bp %>% ly_hist( x = carat, data = diamonds, breaks = 2 )
# density on diamonds
bp %>% ly_density( x = carat, data = diamonds )
# quantile on diamonds
bp %>% ly_quantile(price,group = "color", diamonds)
bp %>% ly_quantile(price,group = "color", diamonds, distn=qnorm)
#demo a transform
bp %>%
ly_points( cyl, mpg^2, mtcars ) %>%
# not transformed
ly_points( cyl, mpg, mtcars, color = "red" ) %>%
# axis need to come after layers specified
y_axis( log = T )
# set vs map color
bp %>%
ly_points( mpg, wt, mtcars, color = "purple")
bp %>%
ly_points( mpg, wt, data.frame(name=rownames(mtcars),mtcars), color = cyl, hover = list(name))
# boxplot
data("Oxboys", package = "nlme")
bp %>%
ly_boxplot( Occasion, height, Oxboys )

rbokeh example
library(rbokeh)
library(ggplot2)
data(diamonds)
bp <- figure( height = 400, width = 700 )
bp %>% ly_points(carat,price,diamonds,color=cut,size=5)
# add hover
bp %>%
ly_points(carat,price,diamonds,color=cut,size=1,hover=list(cut,clarity,color))
# make it a hexbin
bp %>%
ly_hexbin( carat, price, diamonds )
# make it a hexbin with facet by color
lapply(
levels(unique(diamonds$color))
,function(c){
figure( height = 300, width = 300, title = paste0("Color: ",c) ) %>%
ly_hexbin( carat, log(price), diamonds[which(diamonds$color==c),] )
}
) %>%
grid_plot( nrow = 3, ncol = 3, same_axes = T )
# histogram on diamonds
bp %>% ly_hist( x = carat, data = diamonds, breaks = 2 )
# density on diamonds
bp %>% ly_density( x = carat, data = diamonds )
# quantile on diamonds
bp %>% ly_quantile(price,group = "color", diamonds)
bp %>% ly_quantile(price,group = "color", diamonds, distn=qnorm)
#demo a transform
bp %>%
ly_points( cyl, mpg^2, mtcars ) %>%
# not transformed
ly_points( cyl, mpg, mtcars, color = "red" ) %>%
# axis need to come after layers specified
y_axis( log = T )
# set vs map color
bp %>%
ly_points( mpg, wt, mtcars, color = "purple")
bp %>%
ly_points( mpg, wt, data.frame(name=rownames(mtcars),mtcars), color = cyl, hover = list(name))
# boxplot
data("Oxboys", package = "nlme")
bp %>%
ly_boxplot( Occasion, height, Oxboys )

rbokeh example
library(rbokeh)
library(ggplot2)
data(diamonds)
bp <- figure( height = 400, width = 700 )
bp %>% ly_points(carat,price,diamonds,color=cut,size=5)
# add hover
bp %>%
ly_points(carat,price,diamonds,color=cut,size=1,hover=list(cut,clarity,color))
# make it a hexbin
bp %>%
ly_hexbin( carat, price, diamonds )
# make it a hexbin with facet by color
lapply(
levels(unique(diamonds$color))
,function(c){
figure( height = 300, width = 300, title = paste0("Color: ",c) ) %>%
ly_hexbin( carat, log(price), diamonds[which(diamonds$color==c),] )
}
) %>%
grid_plot( nrow = 3, ncol = 3, same_axes = T )
# histogram on diamonds
bp %>% ly_hist( x = carat, data = diamonds, breaks = 2 )
# density on diamonds
bp %>% ly_density( x = carat, data = diamonds )
# quantile on diamonds
bp %>% ly_quantile(price,group = "color", diamonds)
bp %>% ly_quantile(price,group = "color", diamonds, distn=qnorm)
#demo a transform
bp %>%
ly_points( cyl, mpg^2, mtcars ) %>%
# not transformed
ly_points( cyl, mpg, mtcars, color = "red" ) %>%
# axis need to come after layers specified
y_axis( log = T )
# set vs map color
bp %>%
ly_points( mpg, wt, mtcars, color = "purple")
bp %>%
ly_points( mpg, wt, data.frame(name=rownames(mtcars),mtcars), color = cyl, hover = list(name))
# boxplot
data("Oxboys", package = "nlme")
bp %>%
ly_boxplot( Occasion, height, Oxboys )
ggplot2
p <- ggplot(diamonds, aes(x=factor(color), y=carat))
>
> # Boxplot of diamond carat as a function of diamond color
> p + geom_boxplot()
p + geom_boxplot() + coord_flip()
>
> # Plot3: Set aesthetics to fixed value
> p + geom_boxplot(fill = "palegreen", color = "blue4", size=0.5, outlier.color = "blue4", outlier.size = 2)
>
> # Plot4: Vary fill by diamond color
> p + geom_boxplot(aes(fill=factor(color)))
>
> # Plot5: Add more dimensions with new aesthetic mappings
> p + geom_boxplot(aes(fill = factor(cut))

plotly
> library(plotly)
>
> p <- ggplot(mtcars, aes(x = factor(gear), y = mpg, color = cyl)) +
+ geom_boxplot() +
+ geom_jitter(size = 5)
>
>
> ggplotly(p)

ggplot2
> library(ggplot2)
> ggplot(diamonds, aes(cut, color)) + geom_jitter(aes(color = cut), size = 0.5)

ggplot2
df <- ToothGrowth
> df$dose <- as.factor(df$dose)
> data_summary <- function(data, varname, grps){
+ require(plyr)
+ summary_func <- function(x, col){
+ c(mean = mean(x[[col]], na.rm=TRUE),
+ sd = sd(x[[col]], na.rm=TRUE))
+ }
+ data_sum<-ddply(data, grps, .fun=summary_func, varname)
+ data_sum <- rename(data_sum, c("mean" = varname))
+ return(data_sum)
+ }
> df2 <- data_summary(df, varname="len", grps= "dose")
Loading required package: plyr
Attaching package: ‘plyr’
The following objects are masked from ‘package:plotly’:
arrange, mutate, rename, summarise
The following object is masked from ‘package:network’:
is.discrete
The following object is masked from ‘package:graph’:
join
Warning message:
package ‘plyr’ was built under R version 3.3.1
> # Convert dose to a factor variable
> df2$dose=as.factor(df2$dose)
> head(df2)
dose len sd
1 0.5 10.605 4.499763
2 1 19.735 4.415436
3 2 26.100 3.774150
> f <- ggplot(df2, aes(x = dose, y = len,
+ ymin = len-sd, ymax = len+sd))
> f + geom_crossbar()
> # color by groups
> f + geom_crossbar(aes(color = dose))
> # Change color manually
> f + geom_crossbar(aes(color = dose)) +
+ scale_color_manual(values = c("#999999", "#E69F00", "#56B4E9"))+
+ theme_minimal()
> # fill by groups and change color manually
> f + geom_crossbar(aes(fill = dose)) +
+ scale_fill_manual(values = c("#999999", "#E69F00", "#56B4E9"))+
+ theme_minimal() + ggtitle("prepared by Volkan OBAN in R")
> library(ggthemes)
> f + geom_crossbar()
> # color by groups
> f + geom_crossbar(aes(color = dose))
> # Change color manually
> f + geom_crossbar(aes(color = dose)) +
+ scale_color_manual(values = c("#999999", "#E69F00", "#56B4E9"))+
+ theme_minimal()
> # fill by groups and change color manually
> f + geom_crossbar(aes(fill = dose)) +
+ scale_fill_manual(values = c("#999999", "#E69F00", "#56B4E9"))+ theme_economist() + scale_colour_economist()

ggplot2
ggplot(diamonds, aes(cut, color)) + ggtitle("prepared in R by Volkan OBAN \n ggplot2 package-data=diamonds") +
+ geom_jitter(aes(color = cut), size = 0.5

lattice package in R
x<- c(-1,-0.75,-0.5,-0.25,0,0.25,0.5,0.75,1)
> y<- c(-1,-0.75,-0.5,-0.25,0,0.25,0.5,0.75,1)
> z<- c(0.226598762, 0.132395904, 0.14051906, 0.208607098, 0.320840304,
+ 0.429423216, 0.54086732, 0.647792527, 0.256692375,
+ 0.256403273, 0.172881269, 0.121978079, 0.156718831, 0.17175081,
+ 0.32791861, 0.420194456, 0.493195109, 0.619020921,
+ 0.278066455, 0.199822296, 0.140827896, 0.140139205, 0.206984231,
+ 0.2684947, 0.340728872, 0.422645622, 0.501908648,
+ 0.285697424, 0.22749307, 0.16881002, 0.13354722, 0.149532449,
+ 0.213353293, 0.283777474, 0.355946993, 0.427175997,
+ 0.294521663, 0.236133131, 0.18710497, 0.14828074, 0.145457711,
+ 0.182992988, 0.228281887, 0.291865148, 0.341808458,
+ 0.271987072, 0.252962505, 0.201123092, 0.162942848, 0.14828074,
+ 0.167205292, 0.214481881, 0.27141981, 0.332162403,
+ 0.268966875, 0.253628745, 0.213509108, 0.180342353, 0.151623426,
+ 0.1617176, 0.192572929, 0.243404723, 0.301780548,
+ 0.284462825, 0.25473406, 0.215401758, 0.202840815, 0.171061666,
+ 0.160368388, 0.183680312, 0.226156887, 0.272598273,
+ 0.305655289, 0.247134344, 0.235118253, 0.214725129, 0.185684599,
+ 0.167917048, 0.184066896, 0.218763431, 0.256692375)
>
> model<-data.frame(x,y,z)
z1 <- matrix(z, 9, 9)
> persp(x, y, z1)
> library(lattice)
> g <- expand.grid(x = x, y = y)
> g$z <- z
> wireframe(z ~ x * y, data = g)
> library(lattice)
> g <- expand.grid(x = x, y = y)
> g$z <- z
> wireframe(z ~ x * y,main="Volkan OBAN",data = g)
> library(lattice)
> g <- expand.grid(x = x, y = y)
> g$z <- z
> wireframe(z ~ x * y,data = g)
>

GGally
data(flea)
ggscatmat(flea, columns = 2:4, color="species", alpha=0.8)

GGally-ggplot2
> library(ggplot2)
> pm <- ggpairs(tips, mapping = aes(color = sex), columns = c("total_bill", "time", "tip"))
> pm

GGally
>
> data(tips, package = "reshape")
> pm <- ggpairs(tips)
> pm

GGally-ggduo
swiss <- datasets::swiss
# add a 'fake' column
swiss$Residual <- seq_len(nrow(swiss))
# calculate all residuals prior to display
residuals <- lapply(swiss[2:6], function(x) {
summary(lm(Fertility ~ x, data = swiss))$residuals
})
# calculate a consistent y range for all residuals
y_range <- range(unlist(residuals))
# custom function to display continuous data. If the y variable is "Residual", do custom work.
lm_or_resid <- function(data, mapping, ..., line_color = "red", line_size = 1) {
if (as.character(mapping$y) != "Residual") {
return(ggally_smooth_lm(data, mapping, ...))
}
# make residual data to display
resid_data <- data.frame(
x = data[[as.character(mapping$x)]],
y = residuals[[as.character(mapping$x)]]
)
ggplot(data = data, mapping = mapping) +
geom_hline(yintercept = 0, color = line_color, size = line_size) +
ylim(y_range) +
geom_point(data = resid_data, mapping = aes(x = x, y = y), ...)
}
# plot the data
ggduo(
swiss,
2:6, c(1,7),
types = list(continuous = lm_or_resid)
)

GGally-ggduo
> library(GGally)
> psych <- read.csv("http://www.ats.ucla.edu/stat/data/mmreg.csv")
> colnames(psych) <- c("Control", "Concept", "Motivation", "Read", "Write", "Math", "Science", "Sex")
> psych <- data.frame(
+ Motivation = psych$Motivation,
+ Self.Concept = psych$Concept,
+ Locus.of.Control = psych$Control,
+ Read = psych$Read,
+ Write = psych$Write,
+ Math = psych$Math,
+ Science = psych$Science,
+ Sex = c("0" = "Male", "1" = "Female")[as.character(psych$Sex)]
+ )
> ggduo(
+ psych, 1:3, 4:8,
+ types = list(continuous = "smooth_lm"),
+ title = "Between Academic and Psychological Variable Correlation",
+ xlab = "Psychological",
+ ylab = "Academic"
+ )

maps-ggmap-mapproj
map = suppressMessages(get_map(location = 'Turkey', zoom = 4))
> ggmap(map)

maps-ggmap-mapproj
> suppressMessages(library(maps))
> suppressMessages(library(ggmap))
> suppressMessages(library(mapproj)
map1 = suppressMessages(get_map(
+ location = 'Maslak', zoom = 14, #zoom-in level
+ maptype="satellite")) #map type
> ggmap(map1)

GGally
> data(tips, package = "reshape")
> plotList <- list(
+ qplot(total_bill, tip, data = subset(tips, smoker == "No" & sex == "Female")) +
+ facet_grid(time ~ day),
+ qplot(total_bill, tip, data = subset(tips, smoker == "Yes" & sex == "Female")) +
+ facet_grid(time ~ day),
+ qplot(total_bill, tip, data = subset(tips, smoker == "No" & sex == "Male")) +
+ facet_grid(time ~ day),
+ qplot(total_bill, tip, data = subset(tips, smoker == "Yes" & sex == "Male")) +
+ facet_grid(time ~ day)
pm <- ggmatrix(
plotList, nrow = 2, ncol = 2,
yAxisLabels = c("Female", "Male"),
xAxisLabels = c("Non Smoker", "Smoker"),
title = "Total Bill vs Tip",
showStrips = NULL # default
)
pm

GGally
> library(GGally)
> psych <- read.csv("http://www.ats.ucla.edu/stat/data/mmreg.csv")
> colnames(psych) <- c("Control", "Concept", "Motivation", "Read", "Write", "Math", "Science", "Sex")
> psych <- data.frame(
+ Motivation = psych$Motivation,
+ Self.Concept = psych$Concept,
+ Locus.of.Control = psych$Control,
+ Read = psych$Read,
+ Write = psych$Write,
+ Math = psych$Math,
+ Science = psych$Science,
+ Sex = c("0" = "Male", "1" = "Female")[as.character(psych$Sex)]
+ )
> ggpairs(psych, 4:8, title = "prepared by Volkan OBAN using R-GGally pack \n Within Academic Variables")

GGally
> library(GGally)
> psych <- read.csv("http://www.ats.ucla.edu/stat/data/mmreg.csv")
> colnames(psych) <- c("Control", "Concept", "Motivation", "Read", "Write", "Math", "Science", "Sex")
> psych <- data.frame(
+ Motivation = psych$Motivation,
+ Self.Concept = psych$Concept,
+ Locus.of.Control = psych$Control,
+ Read = psych$Read,
+ Write = psych$Write,
+ Math = psych$Math,
+ Science = psych$Science,
+ Sex = c("0" = "Male", "1" = "Female")[as.character(psych$Sex)]
+ )
> ggpairs(psych, 1:3, title = " Within Psychological Variables")

sjPlot and sjmisc package
data(efc)
> # Function call when label attributes are attached
> sjp.xtab(efc$e42dep, efc$e16sex)
> sjp.xtab(efc$e42dep, efc$e16sex, title="prepared in R by Volkan OBAN")

sjPlot and sjmisc package
library(sjPlot)
> library(sjmisc)
> # init default theme for plots
> sjp.setTheme(geom.label.size = 2.5, axis.title.size = .9, axis.textsize = .9)
sjp.frq(dummy, title = "prepared by VOLKAN OBAN using R")
> dummy <- set_labels(dummy, c("very low", "low", "mid", "hi"))
> dummy <- set_label(dummy, "Humidity")
> # check structure of dummy
> str(dummy)
atomic [1:200] 2 2 3 4 3 3 2 3 2 3 ...
- attr(*, "labels")= Named num [1:4] 1 2 3 4
..- attr(*, "names")= chr [1:4] "very low" "low" "mid" "hi"
- attr(*, "label")= chr "Humidity"
> sjp.frq(dummy, title = "prepared by VOLKAN OBAN using R")
>

pair plot
data(iris)
> pairs(iris[1:5], main = "Iris Data", pch = 21, bg = c("red", "green3", "blue")[unclass(iris$Species)])

ggplot2
> data(mpg)
> g<-ggplot(mpg, aes(displ, hwy, color=factor(year)))
>
> g+geom_point()
> g+geom_point()+facet_grid(drv~cyl, margins=TRUE)
ggplot2
options(repr.plot.width = 8)
> options(repr.plot.height = 6)
>
> ggplot(clim, aes(Year, CO2))+geom_line(color='black')+geom_point(color='red') +ggtitle("Carbondioxide Concentration")+xlab('prepared in R-ggplot2 by VOLKAN OBAN')+ylab('ppm')+
+ geom_vline(xintercept = c(1990,2000),colour="green", linetype = "longdash")
>

ggplot2 example
> clim<-read.csv("climate_change.csv")
> options(repr.plot.width = 8)
> options(repr.plot.height = 6)
>
library(ggplot2)
> ggplot(clim, aes(Year, Temp))+geom_line(color='green')+geom_point()+ggtitle('Temperature Change')+xlab('prepared in R-ggplot2 by VOLKAN OBAN')+ylab('Temperature')+stat_smooth(colour='blue', span=0.2)

ggplot2
dsub <- diamonds[ sample(nrow(diamonds), 1000), ]
ggplot(dsub, aes(x = cut, y = carat, fill = clarity)) +
+ geom_boxplot(outlier.size = 0) +
+ geom_point(pch = 21, position = position_jitterdodge())

ggplot2 example
> p <- ggplot(ToothGrowth, aes(x=factor(dose), y=len, fill=supp)) +
+ scale_fill_manual(values=c("#FF0000", "white")) + ggtitle("prepared in R by Volkan OBAN ")
> p + geom_dotplot(binaxis="y", position="dodge", stackdir="center", binwidth=2)

geom_boxplot
g <- ggplot(dt, aes_string(x="Group", y="Duration")) + coord_flip() +
geom_boxplot(aes(ymin=..lower.., ymax=..upper..), fatten=1.1, lwd=.1, outlier.shape=NA) +
geom_dotplot(data=dt[EndType==1], aes(fill=EndType), fill="black", binaxis="y", stackdir="up", method="histodot", binwidth=15, dotsize=.5) + geom_dotplot(data=dt[EndType==0], aes(fill=EndType), fill="white", binaxis="y", stackdir="down", method="histodot", binwidth=15, dotsize=.5)
> print(g)

ggplot2 - Creating basic Jitter
> # Creating basic Jitter
> ggplot(ChickWeight, aes(x = Diet, y = weight)) +
+ geom_boxplot(notch = TRUE) +
+ geom_jitter(position = position_jitter(0.5), aes(colour = Diet)) + ggtitle("prepared in R by Volkan OBAN \n ggplot Density Plot \n data(ChickWeight) ")
ggplot2 example
ggplot(data = diamonds, aes(x = price, fill = cut)) +
geom_histogram(binwidth = 250, color = "black")+ ggtitle("prepared in R by Volkan OBAN \n ggplot Density Plot \n data(diamonds) ")
ggplot2 example
ggplot(data = diamonds, aes(x = price, fill = cut)) +
geom_histogram(binwidth = 250, color = "midnightblue") +
theme(legend.position = "top")

ggplot2 example
> ggplot(data = diamonds, aes(x = price, fill = cut))
+ geom_density(adjust = 1/5, color = "midnightblue")
+ facet_wrap(~ cut, scale = "free")
+ labs(title="GGPLOT Density Plot", x="Price in Dollars", y="Density")
+ theme_dark()

ggplot2 example
ggplot(data = diamonds, aes(x = price, fill = cut)) +
geom_density(adjust = 1/5, color = "midnightblue") +
theme(legend.position = "top")

geom_dotplot
library(ggplot2)
>
> # Create a Dot plot
> ggplot(airquality, aes(x = Wind, fill = factor(Month))) +
+ geom_dotplot(binwidth = 1.5)

ggplot2 - geom_dotplot
library(ggplot2)
>
> # Create a Dot plot
> ggplot(airquality, aes(x = factor(Month), fill = factor(Month),
+ y = Wind)) +
+ geom_dotplot(binaxis = "y", stackdir = "center",
+ color = "gold") + ggtitle("prepared in R by Volkan OBAN \n ggplot2 - geom_dotplot")

ggplot2 example
ggplot(diamonds, aes(carat, depth)) +
geom_boxplot(aes(group = plyr::round_any(carat, 0.1))) +
xlim(NA, 2.05) + ggtitle("prepared by Volkan OBAN using R-ggplot2")

ggplot2 example
ggplot(mpg, aes(drv, displ, fill = drv)) +
geom_dotplot(binaxis = "y", stackdir = "center") + ggtitle("prepared by Volkan OBAN using R-ggplot2")

ggplot2
ggplot(diamonds, aes(carat, depth)) +
geom_boxplot(aes(group = plyr::round_any(carat, 0.1))) +
xlim(NA, 2.05)

ggplot2
> data(Oxboys, package = "nlme")
> head(Oxboys)
library(ggplot2)
ggplot(Oxboys, aes(Occasion, height)) +
geom_boxplot() +
geom_line(aes(group = Subject), colour = "#3366FF", alpha = 0.5)
plotly
library(plotly)
> p <- plot_ly(midwest, x = ~percollege, color = ~state, type = "box") %>% layout( title = "prepared in R-plotly by VOLKAN OBAN")
> p

colorful histogram
t<- c(97,93,91,87,86,85,80,78,69,68,67,65,63,62,59,59,54,51,48,45,43,43,38,33,30,27,25,20,18,15,12,7,3,3,3)
> hist(t, main=" colorful histogram \n Distribution of Player Ratings",xlim = c(0,99), breaks=c(seq(2,99,2)), col = c("darkred", "deepskyblue3", "red", "purple","mediumorchid1","darkorange2","black","navyblue"))
manhattanly
> library(manhattanly)
> manhattanly(HapMap,
+ snp = "SNP", gene = "GENE", title="prepared in R-manhattanly pack. by Volkan OBAN \n Manhattan Plot",
+ annotation1 = "ZSCORE", annotation2 = "EFFECTSIZE",
+ highlight = significantSNP)

plotly
p <- plot_ly(y = ~rnorm(50), type = "box") %>%
+ add_trace(y = ~rnorm(50, 1)) %>%
+ add_trace(y=~runif(numcases,min,max+1)) %>%
+ add_trace(y = ~rnorm(80, 1)) %>%
+ add_trace(y = ~rnorm(100, 1)) %>%
+ add_trace(y = ~rnorm(78, 1)) %>%
+ add_trace(y = ~rnorm(98, 1)) %>%
+ add_trace(y = ~rnorm(45, 1)) %>%
+ add_trace(y = ~rnorm(95, 1)) %>%
+ add_trace(y = ~qbinom(0.2, 10, 1/3)) %>%
+ add_trace(y = ~rnorm(95, 1)) %>%
+ add_trace(y = ~runif(numcases,min,max/2)) %>%
+
+ add_trace(y = ~~runif(numcases,min,max+2) %>%
+ add_trace(y = ~pnorm(27.4, mean=50, sd=20)) %>%
+ add_trace(y = ~runif(numcases,min,max+1)) %>%
+ layout( title = "prepared in R-plotly by VOLKAN OBAN"))
> p

Publish Plot
ggplot2
ggthemes
> data = data.frame(student = c("VOLKAN", "OBAN", "GÜL", "GÜLCE", "SEMRA"),
+ percentile = c(25, 95, 54, 70, 99) )
plot + geom_pointrange(aes(ymin = 0, ymax = 100)) + coord_flip() + ggtitle("prepared in R by VOLKAN OBAN") + theme_economist() + scale_colour_economist()
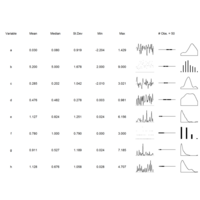
plots/sparktable.R
ref:https://github.com/ebommes/plots/blob/master/sparktable.R
> options(stringsAsFactors = FALSE)
Error: could not find function "ptions"
>
> library(grid)
> library(gtable)
> library(ggplot2)
> library(ggthemes)
>
> randu <- function(n) {
+ r <- sample(1:5, 1)
+ if(r == 1) return(rnorm(n));
+ if(r == 2) return(runif(n));
+ if(r == 3) return(rbinom(n, 10, 0.5));
+ if(r == 4) return(rpois(n, 0.8));
+ if(r == 5) return(rexp(n));
+ }
>
> test_intg <- function(x) {
+ if(class(x) != 'numeric') return(TRUE);
+ perc <- length(unique(x)) / length(x)
+
+ if(perc <= 0.05) {
+ return(TRUE)
+ } else {
+ return(FALSE)
+ }
+ }
>
> sumry <- function(df) {
+ df_mean <- sapply(df, mean)
+ df_median <- sapply(df, median)
+ df_sd <- sapply(df, sd)
+ df_min <- sapply(df, min)
+ df_max <- sapply(df, max)
+ data.frame(Variable = names(df),
+ Mean = format(round(df_mean, 3), nsmall = 3),
+ Median = format(round(df_median, 3), nsmall = 3),
+ St.Dev = format(round(df_sd, 3), nsmall = 3),
+ Min = format(round(df_min, 3), nsmall = 3),
+ Max = format(round(df_max, 3), nsmall = 3))
+ }
>
> theme_spark <- function() {
+ theme_tufte() +
+ theme(axis.title = element_blank(),
+ axis.text.y = element_blank(),
+ axis.ticks = element_blank(),
+ axis.text.x = element_blank())
+ }
>
> tplotter <- function(x) {
+ df_tmp <- data.frame(x = c(1:length(x)), y = x)
+
+ if(test_intg(x) == TRUE) {
+ p <- ggplot(df_tmp, aes(x = x, y = y)) +
+ theme_spark() +
+ geom_hline(yintercept = 0, colour = 'darkgrey') +
+ geom_point(fill = 'black', size = 0.1)
+ } else {
+ p <- ggplot(df_tmp, aes(x = x, y = y)) +
+ theme_spark() +
+ geom_hline(yintercept = 0, colour = 'darkgrey') +
+ geom_line(colour = 'black')
+ }
+
+ return(ggplotGrob(p))
+ }
>
> dplotter <- function(x) {
+ df_tmp <- data.frame(x = c(1:length(x)), y = x)
+
+ if(test_intg(x) == TRUE) {
+ p <- ggplot(df_tmp, aes(x = y)) +
+ theme_spark() +
+ geom_bar(fill = 'black', width = 0.25)
+ } else {
+ p <- ggplot(df_tmp, aes(x = y)) +
+ theme_spark() +
+ geom_density(color = 'black')
+ }
+
+ return(ggplotGrob(p))
+ }
>
> bplotter <- function(x) {
+ df_tmp <- data.frame(x = c(1:length(x)), y = x)
+
+ p <- ggplot(df_tmp, aes(x = y, y = y)) +
+ theme_spark() +
+ coord_flip() + ggtitle("prepared in R by VOLKAN OBAN") +
+ geom_tufteboxplot(median.type = 'line', whisker.type = 'line',
+ hoffset = 0, width = 3, voffset = 0.02)
+
+ return(ggplotGrob(p))
+ }
>
> n <- 50
> set.seed(1234)
> df <- data.frame(a = randu(n), b = randu(n), c = randu(n), d = randu(n),
+ e = randu(n), f = randu(n), g = randu(n), h = randu(n))
>
> df_sumry <- sumry(df)
>
> df.names <- names(df_sumry)
> m <- ncol(df_sumry)
> n <- nrow(df_sumry)
>
> funs <- c('tplotter', 'bplotter', 'dplotter')
>
> gtab <- gtable(unit(rep(1, m + length(funs)), 'null'), unit(rep(1, n + 1), 'null'))
>
> # fill text
> for(i in 1:n) {
+ for(j in 1:ncol(df_sumry)) {
+ if(i == 1) {
+ gtab <- gtable_add_grob(gtab, textGrob(df.names[j]),
+ t = i, l = j, r = j)
+ }
+
+ gtab <- gtable_add_grob(gtab, textGrob(df_sumry[i, j]),
+ t = i + 1, l = j, r = j)
+ }
+
+ for(j in 1:length(funs)) {
+ gtab <- gtable_add_grob(gtab, do.call(funs[j], list(df[, i])),
+ t = i + 1, l = m + j, r = m + j)
+ }
+
+ }
>
> gtab <- gtable_add_grob(gtab, textGrob(paste('# Obs. =', nrow(df))),
+ t = 1, l = m + 1, r = m + length(funs))
>
> dev.new(width = 0.79 * (m + 3), height = 0.42 * n)
NULL
> grid.draw(gtab)
plotly example
count <- 3000
>
> x <- c()
> y <- c()
> z <- c()
> c <- c()
>
> for (i in 1:count) {
+ r <- i * (count - i)
+ x <- c(x, r * cos(i / 30))
+ y <- c(y, r * sin(i / 30))
+ z <- c(z, i)
+ c <- c(c, i)
+ }
>
> data <- data.frame(x, y, z, c)
>
> p <- plot_ly(data, x = ~x, y = ~y, z = ~z, type = 'scatter3d', mode = 'lines',
+ line = list(width = 4, color = ~c, colorscale = list(c(0,'#BA52ED'), c(1,'#FCB040'))))
>
> p

data visulazition in R an example
library("maps")
require(ggplot2)
library(ggsubplot)
world.map <- map("world", plot = FALSE, fill = TRUE)
world_map <- map_data("world")
require(lattice)
require(latticeExtra)
# Calculate the mean longitude and latitude per region (places where subplots are plotted)
library(plyr)
cntr <- ddply(world_map,.(region),summarize,long=mean(long),lat=mean(lat))
# example data
myd <- data.frame (region = c("USA","China","USSR","Brazil", "Australia","India", "Nepal", "Canada",
"South Africa", "South Korea", "Philippines", "Mexico", "Finland",
"Egypt", "Chile", "Greenland"),
frequency = c(501, 350, 233, 40, 350, 150, 180, 430, 233, 120, 96, 87, 340, 83, 99, 89))
subsetcntr <- subset(cntr, region %in% c("USA","China","USSR","Brazil", "Australia","India", "Nepal", "Canada",
"South Africa", "South Korea", "Philippines", "Mexico", "Finland",
"Egypt", "Chile", "Greenland"))
simdat <- merge(subsetcntr, myd)
colnames(simdat) <- c( "region","long","lat", "myvar" )
panel.3dmap <- function(..., rot.mat, distance, xlim,
ylim, zlim, xlim.scaled, ylim.scaled, zlim.scaled) {
scaled.val <- function(x, original, scaled) {
scaled[1] + (x - original[1]) * diff(scaled)/diff(original)
}
m <- ltransform3dto3d(rbind(scaled.val(world.map$x,
xlim, xlim.scaled), scaled.val(world.map$y, ylim,
ylim.scaled), zlim.scaled[1]), rot.mat, distance)
panel.lines(m[1, ], m[2, ], col = "green4")
}
p2 <- cloud(myvar ~ long + lat, simdat, panel.3d.cloud = function(...) {
panel.3dmap(...)
panel.3dscatter(...)
}, type = "h", col = "purple", scales = list(draw = FALSE), zoom = 1.1,
xlim = world.map$range[1:2], ylim = world.map$range[3:4],
xlab = NULL, ylab = NULL, zlab = NULL, aspect = c(diff(world.map$range[3:4])/diff(world.map$range[1:2]),
0.3), panel.aspect = 0.75, lwd = 2, screen = list(z = 30,
x = -60), par.settings = list(axis.line = list(col = "transparent"),
box.3d = list(col = "transparent", alpha = 0)))
p2
mapview
m <- leaflet() %>% addTiles()
garnishMap(m, "addMouseCoordinates") # same as
garnishMap(m, addMouseCoordinates)
## add more than one with named argument
library(raster)
m1 <- garnishMap(m, addMouseCoordinates, mapview:::addHomeButton,
ext = extent(breweries91))
m1
## even more flexible
m2 <- garnishMap(m1, addCircleMarkers, data = breweries91)
garnishMap(m2, addPolygons, data = gadmCHE, popup = popupTable(gadmCHE),
fillOpacity = 0.8, color = "black", fillColor = "#BEBEBE")
mapview
m <- leaflet() %>% addTiles()
garnishMap(m, "addMouseCoordinates") # same as
garnishMap(m, addMouseCoordinates)
## add more than one with named argument
library(raster)
m1 <- garnishMap(m, addMouseCoordinates, mapview:::addHomeButton,
ext = extent(breweries91))
m1
## even more flexible
m2 <- garnishMap(m1, addCircleMarkers, data = breweries91)
garnishMap(m2, addPolygons, data = gadmCHE, popup = popupTable(gadmCHE),
fillOpacity = 0.8, color = "black", fillColor = "#BEBEBE")

wordcloud2-E=m.c^2
wordcloud2(demoFreq, figPath = "em.png", size = 1.5, color = "white", backgroundColor="black")

ggplot2 Time Series Heatmaps
require(quantmod)
require(ggplot2)
require(reshape2)
require(plyr)
require(scales)
# Download some Data, e.g. the CBOE VIX
getSymbols("^VIX",src="yahoo")
# Make a dataframe
dat<-data.frame(date=index(VIX),VIX)
# We will facet by year ~ month, and each subgraph will
# show week-of-month versus weekday
# the year is simple
dat$year<-as.numeric(as.POSIXlt(dat$date)$year+1900)
# the month too
dat$month<-as.numeric(as.POSIXlt(dat$date)$mon+1)
# but turn months into ordered facors to control the appearance/ordering in the presentation
dat$monthf<-factor(dat$month,levels=as.character(1:12),labels=c("Jan","Feb","Mar","Apr","May","Jun","Jul","Aug","Sep","Oct","Nov","Dec"),ordered=TRUE)
# the day of week is again easily found
dat$weekday = as.POSIXlt(dat$date)$wday
# again turn into factors to control appearance/abbreviation and ordering
# I use the reverse function rev here to order the week top down in the graph
# you can cut it out to reverse week order
dat$weekdayf<-factor(dat$weekday,levels=rev(1:7),labels=rev(c("Mon","Tue","Wed","Thu","Fri","Sat","Sun")),ordered=TRUE)
# the monthweek part is a bit trickier
# first a factor which cuts the data into month chunks
dat$yearmonth<-as.yearmon(dat$date)
dat$yearmonthf<-factor(dat$yearmonth)
# then find the "week of year" for each day
dat$week <- as.numeric(format(dat$date,"%W"))
# and now for each monthblock we normalize the week to start at 1
dat<-ddply(dat,.(yearmonthf),transform,monthweek=1+week-min(week))
# Now for the plot
P<- ggplot(dat, aes(monthweek, weekdayf, fill = VIX.Close)) +
geom_tile(colour = "white") + facet_grid(year~monthf) + scale_fill_gradient(low="red", high="yellow") +
options(title = "Time-Series Calendar Heatmap") + xlab("Week of Month") + ylab("")
P

library(dplyr) library(tidyr) library(rvest) library(rcdimple)
library(dplyr)
library(tidyr)
library(rvest)
library(rcdimple)
# Get the table from the Census database with rvest
url <- "http://www.census.gov/population/international/data/idb/region.php?N=%20Results%20&T=10&A=separate&RT=0&Y=2015,2020,2025,2030,2035,2040,2045,2050&R=-1&C=IN"
df <- url %>%
html() %>%
html_nodes("table") %>%
html_table() %>%
data.frame()
names(df) <- c("Year", "Age", "total", "Male", "Female", "percent", "pctMale", "pctFemale", "sexratio")
cols <- c(1, 3:9)
df[,cols] <- apply(df[,cols], 2, function(x) as.numeric(as.character(gsub(",", "", x))))
# Format the table with dplyr and tidyr
df1 <- df %>%
mutate(Order = 1:nrow(df),
Male = -1 * Male) %>%
filter(Age != "Total") %>%
select(Year, Age, Male, Female, Order) %>%
gather(Gender, Population, -Age, -Order, -Year)
max_x <- plyr::round_any(max(df1$Population), 10000, f = ceiling)
min_x <- plyr::round_any(min(df1$Population), 10000, f = floor)
# Build the chart with rcdimple
df1 %>%
dimple(x = "Population", y = "Age", group = "Gender", type = 'bar', storyboard = "Year") %>%
yAxis(type = "addCategoryAxis", orderRule = "Order") %>%
xAxis(type = "addMeasureAxis", overrideMax = max_x, overrideMin = min_x) %>%
default_colors(c("green", "orange")) %>%
add_legend() %>%
add_title(html = "<h3 style='font-family:Helvetica; text-align: center;'>India's population, 2015-2050</h3>") %>%
# Here, I'll pass in some JS code to make all the values on the X-axis and in the tooltip absolute values
tack(., options = list(
chart = htmlwidgets::JS("
function(){
var self = this;
// x axis should be first or [0] but filter to make sure
self.axes.filter(function(ax){
return ax.position == 'x'
})[0] // now we have our x axis set _getFormat as before
._getFormat = function () {
return function(d) {
return d3.format(',.0f')(Math.abs(d) / 1000000) + 'm';
};
};
// return self to return our chart
return self;
}
")) )

rcharts_pyramids
source('https://raw.githubusercontent.com/walkerke/teaching-with-datavis/master/pyramids/rcharts_pyramids.R')
d> library(rCharts)
d> nPyramid('QA', 2014, colors = c('darkred', 'silver'))
d> nPyramid('QA', 2014,colors = c('darkred', 'silver'))
geom_ribbon() function in ggplot2.
huron <- data.frame(year = 1875:1972, level = as.vector(LakeHuron))
h <- ggplot(huron, aes(year))
h + geom_ribbon(aes(ymin=0, ymax=level))
h +
geom_ribbon(aes(ymin = level - 1, ymax = level + 1), fill = "grey70") +
geom_line(aes(y = level)
vcd-perturb packages.
> # Baseball data example, from Friendly & Kwan (2009)
> if (require(vcd) && require(perturb)) {
+ # model, with transformed variables
+ Baseball$logsal <- log(Baseball$sal87)
+ Baseball$years7 <- pmin(Baseball$years,7)
+ base.mod <- lm(logsal ~ years+atbat+hits+homeruns+runs+rbi+walks, data=Baseball)
+ if (require(car)) {
+ # examine variance inflation factors
+ vif(base.mod)
+ }
+ # corresponds to SAS: / collinoint option
+ cd <- colldiag(base.mod, add.intercept=FALSE, center=TRUE)
+ # simplified display
+ print(cd, fuzz=.3)
+ tableplot(cd)
+ }
Zorunlu paket yükleniyor: vcd
Attaching package: ‘vcd’
The following object is masked from ‘package:raster’:
mosaic
Zorunlu paket yükleniyor: perturb
Warning message:
In library(package, lib.loc = lib.loc, character.only = TRUE, logical.return = TRUE, :
there is no package called ‘perturb’
> install.packages("perturb")
Installing package into ‘C:/Users/lenovo/Documents/R/win-library/3.3’
(as ‘lib’ is unspecified)
trying URL 'https://cran.rstudio.com/bin/windows/contrib/3.3/perturb_2.05.zip'
Content type 'application/zip' length 45706 bytes (44 KB)
downloaded 44 KB
package ‘perturb’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:\Users\lenovo\AppData\Local\Temp\Rtmp2hu8MR\downloaded_packages
> library(perturb)
Attaching package: ‘perturb’
The following object is masked from ‘package:raster’:
reclassify
> # Baseball data example, from Friendly & Kwan (2009)
> if (require(vcd) && require(perturb)) {
+ # model, with transformed variables
+ Baseball$logsal <- log(Baseball$sal87)
+ Baseball$years7 <- pmin(Baseball$years,7)
+ base.mod <- lm(logsal ~ years+atbat+hits+homeruns+runs+rbi+walks, data=Baseball)
+ if (require(car)) {
+ # examine variance inflation factors
+ vif(base.mod)
+ }
+ # corresponds to SAS: / collinoint option
+ cd <- colldiag(base.mod, add.intercept=FALSE, center=TRUE)
+ # simplified display
+ print(cd, fuzz=.3)
+ tableplot(cd)
+ }
ggplot
df <- data.frame(x = rnorm(15000),y=rnorm(15000))
ggplot(df,aes(x=x,y=y)) + geom_point() + geom_density2d()

library(cartography)
library(cartography)
# Load data
data(nuts2006)
# set margins
opar <- par(mar = c(0,0,1.2,0))
# Compute the compound annual growth rate
nuts2.df$cagr <- (((nuts2.df$pop2008 / nuts2.df$pop1999)^(1/9)) - 1) * 100
# Plot a layer with the extent of the EU28 countries with only a background color
plot(nuts0.spdf, border = NA, col = NA, bg = "#A6CAE0")
# Plot non european space
plot(world.spdf, col = "#E3DEBF", border=NA, add=TRUE)
# Plot Nuts2 regions
plot(nuts2.spdf, col = "grey60",border = "white", lwd=0.4, add=TRUE)
# Set a custom color palette
cols <- carto.pal(pal1 = "blue.pal", n1 = 2, pal2 = "red.pal", n2 = 4)
# Plot symbols with choropleth coloration
propSymbolsChoroLayer(spdf = nuts2.spdf,
df = nuts2.df,
var = "pop2008", # field in df to plot the symbols sizes
inches = 0.1, # set the symbols sizes
var2 = "cagr", # field in df to plot the colors
col = cols, # symbols colors
breaks = c(-2.43,-1,0,0.5,1,2,3.1), # breaks
border = "grey50", # border colors of the symbols
lwd = 0.75, # symbols width
legend.var.pos = "topright", # legend position
legend.var.values.rnd = -3, # legend value
legend.var.title.txt = "Total Population", # size legend title
legend.var.style = "e", # legend type
legend.var2.pos = "right", # legend position
legend.var2.title.txt = "Compound Annual\nGrowth Rate") # legend title
# layout
layoutLayer(title = "Demographic trends, 1999-2008", coltitle = "black",
sources = "Eurostat, 2011", scale = NULL,
author = "cartography", frame ="", col = NA)
par(opar)
library(chorddiag)
library(chorddiag)
> m <- matrix(c(11975, 5871, 8916, 2868,
+ 1951, 10048, 2060, 6171,
+ 8010, 16145, 8090, 8045,
+ 1013, 990, 940, 6907),
+ byrow = TRUE,
+ nrow = 4, ncol = 4)
> haircolors <- c("black", "blonde", "brown", "red")
> dimnames(m) <- list(have = haircolors,
+ prefer = haircolors)
> m
> groupColors <- c("#000000", "#FFDD89", "#957244", "#F26223")
> chorddiag(m, groupColors = groupColors, groupnamePadding = 40)
>

ade4
data(chats)
chatsw <- data.frame(t(chats))
chatscoa <- dudi.coa(chatsw, scann = FALSE)
par(mfrow = c(2,2))
table.cont(chatsw, abmean.x = TRUE, csi = 2, abline.x = TRUE,
clabel.r = 1.5, clabel.c = 1.5)
table.cont(chatsw, abmean.y = TRUE, csi = 2, abline.y = TRUE,
clabel.r = 1.5, clabel.c = 1.5)
table.cont(chatsw, x = chatscoa$c1[,1], y = chatscoa$l1[,1],
abmean.x = TRUE, csi = 2, abline.x = TRUE, clabel.r = 1.5, clabel.c = 1.5)
table.cont(chatsw, x = chatscoa$c1[,1], y = chatscoa$l1[,1],
abmean.y = TRUE, csi = 2, abline.y = TRUE, clabel.r = 1.5, clabel.c = 1.5)
par(mfrow = c(1,1))
## Not run:
data(rpjdl)
w <- data.frame(t(rpjdl$fau))
wcoa <- dudi.coa(w, scann = FALSE)
table.cont(w, abmean.y = TRUE, x = wcoa$c1[,1], y = rank(wcoa$l1[,1]),
csi = 0.2, clabel.c = 0, row.labels = rpjdl$lalab, clabel.r = 0.75)

plotrix
library(plotrix)
testdf<-data.frame(Before=c(10,7,5,9),During=c(8,6,2,5),After=c(5,3,4,3))
rownames(testdf)<-c("Red","Green","Blue","Lightblue")
barp(testdf,main="Test addtable2plot",ylab="Value",
names.arg=colnames(testdf),col=2:5)
# show most of the options
addtable2plot(0.7 ,8,testdf,bty="o",display.rownames=TRUE,hlines=TRUE,
vlines=TRUE,title="The table")

matplot
x <- seq(1, 100, 1)
y <- matrix(20*100, nrow=100, ncol=20)
for (i in 1:20) {
y[, i] <- cumsum(rnorm(100))
}
#Build the table
df <- data.frame(x=x, y=y)
head(df)
#Plot the table
matplot(df[, 1], df[, 2:21], type="l", main="Twenty Random Walks", xlab="x", ylab="y")
grid()

Cohort Analysis with Heatmap in R
library(dplyr)
> library(ggplot2)
> library(reshape2)
>
> #simulating dataset
> cohorts <- data.frame()
> set.seed(10)
> for (i in c(1:100)) {
+ coh <- data.frame(cohort=i,
+ date=c(i:100),
+ week.lt=c(1:(100-i+1)),
+ num=replicate(1, sample(c(1:40), 100-i+1, rep=TRUE)),
+ av=replicate(1, sample(c(5:10), 100-i+1, rep=TRUE)))
+ coh$num[coh$week.lt==1] <- sample(c(90:100), 1, rep=TRUE)
+ ifelse(max(coh$date)>1, coh$num[coh$week.lt==2] <- sample(c(75:90), 1, rep=TRUE), NA)
+ ifelse(max(coh$date)>2, coh$num[coh$week.lt==3] <- sample(c(60:75), 1, rep=TRUE), NA)
+ ifelse(max(coh$date)>3, coh$num[coh$week.lt==4] <- sample(c(40:60), 1, rep=TRUE), NA)
+ ifelse(max(coh$date)>34,
+ {coh$num[coh$date==35] <- sample(c(60:85), 1, rep=TRUE)
+ coh$av[coh$date==35] <- 4},
+ NA)
+ ifelse(max(coh$date)>47,
+ {coh$num[coh$date==48] <- sample(c(60:85), 1, rep=TRUE)
+ coh$av[coh$date==48] <- 4},
+ NA)
+ ifelse(max(coh$date)>86,
+ {coh$num[coh$date==87] <- sample(c(60:85), 1, rep=TRUE)
+ coh$av[coh$date==87] <- 4},
+ NA)
+ ifelse(max(coh$date)>99,
+ {coh$num[coh$date==100] <- sample(c(60:85), 1, rep=TRUE)
+ coh$av[coh$date==100] <- 4},
+ NA)
+ coh$gr.marg <- coh$av*coh$num
+ cohorts <- rbind(cohorts, coh)
+ }
>
> cohorts$cohort <- formatC(cohorts$cohort, width=3, format='d', flag='0')
> cohorts$cohort <- paste('coh:week:', cohorts$cohort, sep='')
> cohorts$date <- formatC(cohorts$date, width=3, format='d', flag='0')
> cohorts$date <- paste('cal_week:', cohorts$date, sep='')
> cohorts$week.lt <- formatC(cohorts$week.lt, width=3, format='d', flag='0')
> cohorts$week.lt <- paste('week:', cohorts$week.lt, sep='')
>
> #calculating CLV to date
> cohorts <- cohorts %>%
+ group_by(cohort) %>%
+ mutate(clv=cumsum(gr.marg)/num[week.lt=='week:001'])
>
> #color palette
> cols <- c("#e7f0fa", "#c9e2f6", "#95cbee", "#0099dc", "#4ab04a", "#ffd73e", "#eec73a", "#e29421", "#e29421", "#f05336", "#ce472e")
>
> #Heatmap based on Number of active customers
> t <- max(cohorts$num)
>
> ggplot(cohorts, aes(y=cohort, x=date, fill=num)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Cohort Activity Heatmap (number of customers who purchased - calendar view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> ggplot(cohorts, aes(y=cohort, x=week.lt, fill=num)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Cohort Activity Heatmap (number of customers who purchased - lifetime view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> # Heatmap based on Gross margin
> t <- max(cohorts$gr.marg)
>
> ggplot(cohorts, aes(y=cohort, x=date, fill=gr.marg)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Heatmap based on Gross margin (calendar view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> ggplot(cohorts, aes(y=cohort, x=week.lt, fill=gr.marg)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Heatmap based on Gross margin (lifetime view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> # Heatmap of per customer gross margin
> t <- max(cohorts$av)
>
> ggplot(cohorts, aes(y=cohort, x=date, fill=av)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("Heatmap based on per customer gross margin (calendar view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> ggplot(cohorts, aes(y=cohort, x=week.lt, fill=av)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Heatmap based on per customer gross margin (lifetime view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> # Heatmap of CLV to date
> t <- max(cohorts$clv)
>
> ggplot(cohorts, aes(y=cohort, x=date, fill=clv)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Heatmap based on CLV to date of customers who ever purchased (calendar view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> ggplot(cohorts, aes(y=cohort, x=week.lt, fill=clv)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Heatmap based on CLV to date of customers who ever purchased (lifetime view)")

Cohort Analysis with Heatmap in R
library(dplyr)
> library(ggplot2)
> library(reshape2)
>
> #simulating dataset
> cohorts <- data.frame()
> set.seed(10)
> for (i in c(1:100)) {
+ coh <- data.frame(cohort=i,
+ date=c(i:100),
+ week.lt=c(1:(100-i+1)),
+ num=replicate(1, sample(c(1:40), 100-i+1, rep=TRUE)),
+ av=replicate(1, sample(c(5:10), 100-i+1, rep=TRUE)))
+ coh$num[coh$week.lt==1] <- sample(c(90:100), 1, rep=TRUE)
+ ifelse(max(coh$date)>1, coh$num[coh$week.lt==2] <- sample(c(75:90), 1, rep=TRUE), NA)
+ ifelse(max(coh$date)>2, coh$num[coh$week.lt==3] <- sample(c(60:75), 1, rep=TRUE), NA)
+ ifelse(max(coh$date)>3, coh$num[coh$week.lt==4] <- sample(c(40:60), 1, rep=TRUE), NA)
+ ifelse(max(coh$date)>34,
+ {coh$num[coh$date==35] <- sample(c(60:85), 1, rep=TRUE)
+ coh$av[coh$date==35] <- 4},
+ NA)
+ ifelse(max(coh$date)>47,
+ {coh$num[coh$date==48] <- sample(c(60:85), 1, rep=TRUE)
+ coh$av[coh$date==48] <- 4},
+ NA)
+ ifelse(max(coh$date)>86,
+ {coh$num[coh$date==87] <- sample(c(60:85), 1, rep=TRUE)
+ coh$av[coh$date==87] <- 4},
+ NA)
+ ifelse(max(coh$date)>99,
+ {coh$num[coh$date==100] <- sample(c(60:85), 1, rep=TRUE)
+ coh$av[coh$date==100] <- 4},
+ NA)
+ coh$gr.marg <- coh$av*coh$num
+ cohorts <- rbind(cohorts, coh)
+ }
>
> cohorts$cohort <- formatC(cohorts$cohort, width=3, format='d', flag='0')
> cohorts$cohort <- paste('coh:week:', cohorts$cohort, sep='')
> cohorts$date <- formatC(cohorts$date, width=3, format='d', flag='0')
> cohorts$date <- paste('cal_week:', cohorts$date, sep='')
> cohorts$week.lt <- formatC(cohorts$week.lt, width=3, format='d', flag='0')
> cohorts$week.lt <- paste('week:', cohorts$week.lt, sep='')
>
> #calculating CLV to date
> cohorts <- cohorts %>%
+ group_by(cohort) %>%
+ mutate(clv=cumsum(gr.marg)/num[week.lt=='week:001'])
>
> #color palette
> cols <- c("#e7f0fa", "#c9e2f6", "#95cbee", "#0099dc", "#4ab04a", "#ffd73e", "#eec73a", "#e29421", "#e29421", "#f05336", "#ce472e")
>
> #Heatmap based on Number of active customers
> t <- max(cohorts$num)
>
> ggplot(cohorts, aes(y=cohort, x=date, fill=num)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Cohort Activity Heatmap (number of customers who purchased - calendar view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> ggplot(cohorts, aes(y=cohort, x=week.lt, fill=num)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Cohort Activity Heatmap (number of customers who purchased - lifetime view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> # Heatmap based on Gross margin
> t <- max(cohorts$gr.marg)
>
> ggplot(cohorts, aes(y=cohort, x=date, fill=gr.marg)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Heatmap based on Gross margin (calendar view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> ggplot(cohorts, aes(y=cohort, x=week.lt, fill=gr.marg)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Heatmap based on Gross margin (lifetime view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> # Heatmap of per customer gross margin
> t <- max(cohorts$av)
>
> ggplot(cohorts, aes(y=cohort, x=date, fill=av)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("Heatmap based on per customer gross margin (calendar view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> ggplot(cohorts, aes(y=cohort, x=week.lt, fill=av)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Heatmap based on per customer gross margin (lifetime view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> # Heatmap of CLV to date
> t <- max(cohorts$clv)
>
> ggplot(cohorts, aes(y=cohort, x=date, fill=clv)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Heatmap based on CLV to date of customers who ever purchased (calendar view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> ggplot(cohorts, aes(y=cohort, x=week.lt, fill=clv)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Heatmap based on CLV to date of customers who ever purchased (lifetime view)")
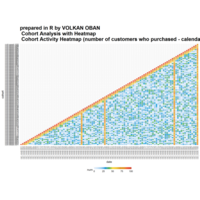
Cohort Analysis with Heatmap in R
library(dplyr)
> library(ggplot2)
> library(reshape2)
>
> #simulating dataset
> cohorts <- data.frame()
> set.seed(10)
> for (i in c(1:100)) {
+ coh <- data.frame(cohort=i,
+ date=c(i:100),
+ week.lt=c(1:(100-i+1)),
+ num=replicate(1, sample(c(1:40), 100-i+1, rep=TRUE)),
+ av=replicate(1, sample(c(5:10), 100-i+1, rep=TRUE)))
+ coh$num[coh$week.lt==1] <- sample(c(90:100), 1, rep=TRUE)
+ ifelse(max(coh$date)>1, coh$num[coh$week.lt==2] <- sample(c(75:90), 1, rep=TRUE), NA)
+ ifelse(max(coh$date)>2, coh$num[coh$week.lt==3] <- sample(c(60:75), 1, rep=TRUE), NA)
+ ifelse(max(coh$date)>3, coh$num[coh$week.lt==4] <- sample(c(40:60), 1, rep=TRUE), NA)
+ ifelse(max(coh$date)>34,
+ {coh$num[coh$date==35] <- sample(c(60:85), 1, rep=TRUE)
+ coh$av[coh$date==35] <- 4},
+ NA)
+ ifelse(max(coh$date)>47,
+ {coh$num[coh$date==48] <- sample(c(60:85), 1, rep=TRUE)
+ coh$av[coh$date==48] <- 4},
+ NA)
+ ifelse(max(coh$date)>86,
+ {coh$num[coh$date==87] <- sample(c(60:85), 1, rep=TRUE)
+ coh$av[coh$date==87] <- 4},
+ NA)
+ ifelse(max(coh$date)>99,
+ {coh$num[coh$date==100] <- sample(c(60:85), 1, rep=TRUE)
+ coh$av[coh$date==100] <- 4},
+ NA)
+ coh$gr.marg <- coh$av*coh$num
+ cohorts <- rbind(cohorts, coh)
+ }
>
> cohorts$cohort <- formatC(cohorts$cohort, width=3, format='d', flag='0')
> cohorts$cohort <- paste('coh:week:', cohorts$cohort, sep='')
> cohorts$date <- formatC(cohorts$date, width=3, format='d', flag='0')
> cohorts$date <- paste('cal_week:', cohorts$date, sep='')
> cohorts$week.lt <- formatC(cohorts$week.lt, width=3, format='d', flag='0')
> cohorts$week.lt <- paste('week:', cohorts$week.lt, sep='')
>
> #calculating CLV to date
> cohorts <- cohorts %>%
+ group_by(cohort) %>%
+ mutate(clv=cumsum(gr.marg)/num[week.lt=='week:001'])
>
> #color palette
> cols <- c("#e7f0fa", "#c9e2f6", "#95cbee", "#0099dc", "#4ab04a", "#ffd73e", "#eec73a", "#e29421", "#e29421", "#f05336", "#ce472e")
>
> #Heatmap based on Number of active customers
> t <- max(cohorts$num)
>
> ggplot(cohorts, aes(y=cohort, x=date, fill=num)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Cohort Activity Heatmap (number of customers who purchased - calendar view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> ggplot(cohorts, aes(y=cohort, x=week.lt, fill=num)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Cohort Activity Heatmap (number of customers who purchased - lifetime view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> # Heatmap based on Gross margin
> t <- max(cohorts$gr.marg)
>
> ggplot(cohorts, aes(y=cohort, x=date, fill=gr.marg)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Heatmap based on Gross margin (calendar view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> ggplot(cohorts, aes(y=cohort, x=week.lt, fill=gr.marg)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Heatmap based on Gross margin (lifetime view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> # Heatmap of per customer gross margin
> t <- max(cohorts$av)
>
> ggplot(cohorts, aes(y=cohort, x=date, fill=av)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("Heatmap based on per customer gross margin (calendar view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> ggplot(cohorts, aes(y=cohort, x=week.lt, fill=av)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Heatmap based on per customer gross margin (lifetime view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> # Heatmap of CLV to date
> t <- max(cohorts$clv)
>
> ggplot(cohorts, aes(y=cohort, x=date, fill=clv)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Heatmap based on CLV to date of customers who ever purchased (calendar view)")
Warning: Ignoring unknown parameters: linewidth
Warning messages:
1: In if (!guide$label) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
2: In if (!guide$ticks) zeroGrob() else { :
the condition has length > 1 and only the first element will be used
>
> ggplot(cohorts, aes(y=cohort, x=week.lt, fill=clv)) +
+ theme_minimal() +
+ geom_tile(colour="white", linewidth=2, width=.9, height=.9) +
+ scale_fill_gradientn(colours=cols, limits=c(0, t),
+ breaks=seq(0, t, by=t/4),
+ labels=c("0", round(t/4*1, 1), round(t/4*2, 1), round(t/4*3, 1), round(t/4*4, 1)),
+ guide=guide_colourbar(ticks=T, nbin=50, barheight=.5, label=T, barwidth=10)) +
+ theme(legend.position='bottom',
+ legend.direction="horizontal",
+ plot.title = element_text(size=20, face="bold", vjust=2),
+ axis.text.x=element_text(size=8, angle=90, hjust=.5, vjust=.5, face="plain")) +
+ ggtitle("prepared in R by VOLKAN OBAN \n Cohort Analysis with Heatmap \n Heatmap based on CLV to date of customers who ever purchased (lifetime view)")

gridExtra-ggplot2
CV_1 <- 0.2
CV_2 <- 0.3
Mean <- 65
sigma_1 <- sqrt(log(1 + CV_1^2))
mu_1 <- log(Mean) - sigma_1^2 / 2
sigma_2 <- sqrt(log(1 + CV_2^2))
mu_2 <- log(Mean) - sigma_2^2 / 2
q <- c(0.25, 0.5, 0.75, 0.9, 0.95)
SummaryTable <- data.frame(
Quantile=paste0(100*q,"%ile"),
Loss_1=round(qlnorm(q, mu_1, sigma_1),1),
Loss_2=round(qlnorm(q, mu_2, sigma_2),1)
)
# Create a plot
library(ggplot2)
plt <- ggplot(data.frame(x=c(20, 150)), aes(x)) +
stat_function(fun=function(x) dlnorm(x, mu_1, sigma_1),
aes(colour="CV_1")) +
stat_function(fun=function(x) dlnorm(x, mu_2, sigma_2),
aes(colour="CV_2")) +
scale_colour_discrete(name = "CV",
labels=c(expression(CV[1]), expression(CV[2]))) +
xlab("Loss") +
ylab("Density") +
ggtitle(paste0("Two log-normal distributions with same mean of ",
Mean,", but different CVs"))
# Create a table plot
library(gridExtra)
names(SummaryTable) <- c("Quantile",
expression(Loss(CV[1])),
expression(Loss(CV[2])))
# Set theme to allow for plotmath expressions
tt <- ttheme_default(colhead=list(fg_params = list(parse=TRUE)))
tbl <- tableGrob(SummaryTable, rows=NULL, theme=tt)
# Plot chart and table into one object
grid.arrange(plt, tbl,
nrow=2,
as.table=TRUE,
heights=c(3,1))

data visulazition in R-Tumblr Likes
> science <- c( 32914, 11566, 4989, 3743, 968, 814, 673, 482, 286, 281 )
> bw <- c( 1694, 1167, 1108, 988, 919, 639, 596, 591, 580, 544 )
> lol <- c( 22627, 18100, 17688, 14374, 13459, 12045, 4711, 3779, 3670, 3393 )
> fashion <- c( 955, 581, 486, 435, 402, 303, 279, 279, 278, 275 )
> architecture <- c( 1426, 461, 433, 251, 230, 219, 194, 194, 175, 167 )
> art <- c( 7492, 2965, 2761, 1316, 544, 435, 413, 331, 307, 296 )
> require(RColorBrewer)
Zorunlu paket yükleniyor: RColorBrewer
> accent = brewer.pal(8, "Accent")
> leg.txt <- c("science", "black & white", "lol", "fashion", "architecture", "art")
> leg.col <- c(accent[1], accent[2], accent[3], accent[4], accent[5], accent[6])
> par(bg="#fafaff")
> plot(science, type="s", log="y", lwd=2, col=accent[1], xlab="x-th most popular blog post", ylab="# likes", main="prepared in R by VOLKAN OBAN \n Distribution of LIKES on tumblr", cex.axis=.8, col.main="#444444", col.axis="#333333", fg="#332211")
> points(bw, type="s", lwd=2, col=accent[2])
> points(lol, type="s", lwd=2, col=accent[3])
> points(fashion, type="s", lwd=3, col=accent[4])
> points(architecture, type="s", lwd=2, col=accent[5])
> points(art, type="s", lwd=2, col=accent[6]) legend("topright", leg.txt, fill=leg.col, title="TAG", text.col="#393939", title.col="#222222", border="#f0ffff", box.col="#666666"

data visulazition in R
library(broom)
library(dplyr)
library(ggplot2)
iris_sub <- select(iris, x1 = Petal.Length, x2 = Petal.Width)
kclusts <- data.frame(k=1:6) %>% group_by(k) %>% do(kclust=kmeans(iris_sub, .$k))
clusters <- kclusts %>% group_by(k) %>% do(tidy(.$kclust[[1]]))
assignments <- kclusts %>% group_by(k) %>% do(augment(.$kclust[[1]], iris_sub))
clusterings <- kclusts %>% group_by(k) %>% do(glance(.$kclust[[1]]))
ggplot(assignments, aes(x = x1, y = x2)) +
facet_wrap(~ k) +
geom_point(aes(color=.cluster)) +
geom_point(data=clusters, size=10, shape="x")

rbokeh
> co2dat <- data.frame(
+ y = co2,
+ x = floor(time(co2)),
+ m = rep(month.abb, 39))
figure(xlim = c(1958, 2010), title="prepared in R-rbokeh by VOLKAN OBAN") %>%
+ ly_lines(x, y, color = m, data = co2dat)

rbokeh example
> wa_cancer <- droplevels(subset(latticeExtra::USCancerRates, state == "Washington"))
> ## y axis sorted by male rate
> ylim <- levels(with(wa_cancer, reorder(county, rate.male)))
>
> figure(ylim = ylim, width = 700,title="prepared in R by Volkan OBAN \n rbokeh package", height = 600, tools = "") %>%
+ ly_segments(LCL95.male, county, UCL95.male,
+ county, data = wa_cancer, color = NULL, width = 2) %>%
+ ly_points(rate.male, county, glyph = 16, data = wa_cancer)

rbokeh example
> figure(xlab="prepared in R by VOLKAN OBAN \n rbokeh package", legend_location = "top_left") %>%
+ ly_quantile(Sepal.Length, group = Species, data = iris)

rbokeh example
> doubles <- read.csv("https://gist.githubusercontent.com/hafen/77f25b556725b3d0066b/raw/10f0e811f09f2b9f0f9ccfb542e296dfac2761d4/doubles.csv")
>
> ly_baseball <- function(x) {
+ base_x <- c(90 * cos(pi/4), 0, 90 * cos(3 * pi/4), 0)
+ base_y <- c(90 * cos(pi/4), sqrt(90^2 + 90^2), 90 * sin(pi/4), 0)
+ distarc_x <- lapply(c(2:4) * 100, function(a)
+ seq(a * cos(3 * pi/4), a * cos(pi/4), length = 200))
+ distarc_y <- lapply(distarc_x, function(x)
+ sqrt((x[1]/cos(3 * pi/4))^2 - x^2))
+
+ x %>%
+ ## boundary
+ ly_segments(c(0, 0), c(0, 0), c(-300, 300), c(300, 300), alpha = 0.4) %>%
+ ## bases
+ ly_crect(base_x, base_y, width = 10, height = 10,
+ angle = 45*pi/180, color = "black", alpha = 0.4) %>%
+ ## infield/outfield boundary
+ ly_curve(60.5 + sqrt(95^2 - x^2),
+ from = base_x[3] - 26, to = base_x[1] + 26, alpha = 0.4) %>%
+ ## distance arcs (ly_arc should work here and would be much simpler but doesn't)
+ ly_multi_line(distarc_x, distarc_y, alpha = 0.4)
+ }
>
> figure(xgrid = FALSE, ygrid = FALSE, width = 630, height = 540,
+ xlab = "Horizontal distance from home plate (ft.) \n prepared by Volkan OBAN using R-rbokeh package",
+ ylab = "Vertical distance from home plate (ft.)") %>%
+ ly_baseball() %>%
+ ly_hexbin(doubles, xbins = 50, shape = 0.77, alpha = 0.75, palette = "Spectral10")

rbokeh example
> p <- figure(width = 800, height = 400,title="prepared by VOLKAN OBAN \n rbokeh packages in R") %>%
+ ly_lines(date, Freq, data = flightfreq, alpha = 0.3) %>%
+ ly_points(date, Freq, data = flightfreq,
+ hover = list(date, Freq, dow), size = 5) %>%
+ ly_abline(v = as.Date("2001-09-11"))
> p

lattice package in R -dotplot
> library(lattice)
> v<-dotplot(reorder(Var2,Freq)~Freq|Var1,data = as.data.frame.table(VADeaths),origin=0,type=c("p","h"),main="R Data Visualization \n lattice package-dotplot",xlab="Number of Deaths per 100 ")
> v

sierpinski triangle-fractal in R.
TurtleGraphics package.
> drawTriangle <- function(points) {
+ turtle_setpos(points[1,1], points[1,2])
+ turtle_goto(points[2,1], points[2,2])
+ turtle_goto(points[3,1], points[3,2])
+ turtle_goto(points[1,1], points[1,2])
+ }
> getMid <- function(p1, p2)
+ (p1+p2)*0.5
> sierpinski <- function(points, degree){
+ drawTriangle(points)
+ if (degree > 0) {
+ p1 <- matrix(c(points[1,], getMid(points[1,], points[2,]),
+ getMid(points[1,], points[3,])), nrow=3, byrow=TRUE)
+ sierpinski(p1, degree-1)
+ p2 <- matrix(c(points[2,], getMid(points[1,], points[2,]),
+ getMid(points[2,], points[3,])), nrow=3, byrow=TRUE)
+ sierpinski(p2, degree-1)
+ p3 <- matrix(c(points[3,], getMid(points[3,], points[2,]),
+ getMid(points[1,], points[3,])), nrow=3, byrow=TRUE)
+ sierpinski(p3, degree-1)
+ }
+ invisible(NULL)
+ }
> turtle_init(520, 500, "clip")
> turtle_do({
+ p <- matrix(c(10, 10, 510, 10, 250, 448), nrow=3, byrow=TRUE)
+ turtle_col("red")
+ sierpinski(p, 6)
+ turtle_setpos(250, 448)
+ })
>

KochSnowflake fractal in R.
TurtleGraphics package
Koch Snowflake
> BMat=rbind(c(0.333,0,0,0.333,-0.333,0),c(0.167,-0.289,0.289,0.167,-0.0830,0.144),c(0.167,0.289,-0.289,0.167,0.083,0.144),c(0.333,0,0,0.333,0.333,0))
>
> # Initial conditions:
> x=0
> y=0
>
> plot(0,0,xlim=c(-0.5,0.5),ylim=c(0,1),col="white",main="prepared in R by VOLKAN OBAN \n Koch Snowflake")
> COLOR=c("green","red","blue","yellow")
>
> for(j in 1:100)
+ {
+ i=sample(1:4,1) # ,prob=c(0.25,0.25,0.25,0.25)
+ x3=x
+ x=BMat[i,1]*x+BMat[i,2]*y+BMat[i,5]
+ y=BMat[i,3]*x3+BMat[i,4]*y+BMat[i,6]
+ points(x,y,pch=".",cex=1, col=COLOR[i])
+ }
> BMat=rbind(c(0.333,0,0,0.333,-0.333,0),c(0.167,-0.289,0.289,0.167,-0.0830,0.144),c(0.167,0.289,-0.289,0.167,0.083,0.144),c(0.333,0,0,0.333,0.333,0))
>
> # Initial conditions:
> x=0
> y=0
>
> plot(0,0,xlim=c(-0.5,0.5),ylim=c(0,1),col="white",main="prepared in R by VOLKAN OBAN \n Koch Snowflake")
> COLOR=c("green","red","blue","yellow")
>
> for(j in 1:2000)
+ {
+ i=sample(1:4,1) # ,prob=c(0.25,0.25,0.25,0.25)
+ x3=x
+ x=BMat[i,1]*x+BMat[i,2]*y+BMat[i,5]
+ y=BMat[i,3]*x3+BMat[i,4]*y+BMat[i,6]
+ points(x,y,pch=".",cex=1, col=COLOR[i])
+ }
>

KochSnowflake
KochSnowflakeExample <- function(){
iterate <- function(T,i){
A = T[ ,1]; B=T[ ,2]; C = T[,3];
if (i == 1){
d = (A + B)/2; h = (C-d); d = d-(1/3)*h;
e = (2/3)*B + (1/3)*A; f = (1/3)*B + (2/3)*A;
}
if (i == 2){
d = B; e = (2/3)*B + (1/3)*C; f = (2/3)*B + (1/3)*A;
}
if (i == 3){
d = (B + C)/2; h = (A-d); d = d-(1/3)*h;
e = (2/3)*C + (1/3)*B; f = (1/3)*C + (2/3)*B;
}
if (i == 4){
d = C; e = (2/3)*C + (1/3)*A; f = (2/3)*C + (1/3)*B;
}
if (i == 5){
d = (A + C)/2; h = (B-d); d = d-(1/3)*h;
e = (2/3)*A + (1/3)*C; f = (1/3)*A + (2/3)*C;
}
if (i == 6){
d = A; e = (2/3)*A + (1/3)*C; f = (2/3)*A + (1/3)*B;
}
if (i == 0){
d = A; e = B; f = C;
}
Tnew = cbind(d,e,f)
return(Tnew); #Return a smaller triangle.
}
draw <- function(T, col=rgb(0,0,0),border=rgb(0,0,0)){
polygon(T[1,],T[2,],col=col,border=border)
}
Iterate = function(T,v,col=rgb(0,0,0),border=rgb(0,0,0)){
for (i in v) T = iterate(T,i);
draw(T,col=col,border=border);
}
#The vertices of the initial triangle:
A = matrix(c(1,0),2,1);
B = matrix(c(cos(2*pi/3), sin(2*pi/3)),2,1);
C = matrix(c(cos(2*pi/3),-sin(2*pi/3)),2,1);
T0 = cbind(A,B,C);
plot(numeric(0),xlim=c(-1.1,1.1),ylim=c(-1.1,1.1),axes=FALSE,frame=FALSE,ann=FALSE);
par(mar=c(0,0,0,0),bg=rgb(1,1,1));
par(usr=c(-1.1,1.1,-1.1,1.1));
#Draw snowflake:
for (i in 0:6) for (j in 0:6) for (k in 0:6) for (l in 0:6) Iterate(T0,c(i,j,k,l));
}
KochSnowflakeExample();

Sierpinski triangle in R.
library(spt)
(abc = st(45,75))
plot(abc, , iter=18)
ggthemes
> library("ggplot2")
> library("ggthemes")
>ggplot(diamonds, aes(price, fill = cut)) +
+ geom_histogram(binwidth = 500) + theme_economist() + scale_colour_economist()
R Data viz.
> nn <- 100
> myData <- data.frame(X = rnorm(nn),
+ Y = rnorm(nn))
> myData$Z <- with(myData, X * Y)
> myData$Y <- myData$Y > 0
>
> # This plot serves only to make a "hard" test case
> # comparing Windows GDI to cairographics.
> zp1 <- ggplot(myData, # \/ Here's a handy little function
+ aes(x = X, fill = cut_number(Z, n = 10))) +ggtitle("prepared in R by VOLKAN OBAN")
> zp1 <- zp1 + geom_abline(aes(intercept = X, slope = X), lwd = 1/5)
> zp1 <- zp1 + geom_density(alpha = 2/3)
> zp1 <- zp1 + theme_bw()
> zp1 <- zp1 + facet_grid(~ Y)
> zp1 <- zp1 + scale_fill_manual(values = colorRampPalette(rev(brewer.pal(11, "Spectral")))(10),
+ guide = "none")
> print(zp1)
>
> ggsave(plot = zp1, "Standard ggsave.png", h = 9/3, w = 16/3)
> ggsave(plot = zp1, "Cairo ggsave.png", h = 9/3, w = 16/3, type = "cairo-png")
beanplot package
library(beanplot)
> beanplot(rnorm(100),rnorm(150), rnorm(180),rnorm(50), runif(85), runif(70),runif(30),col="red", xlab="prepared by VOLKAN OBAN using R-beanplot")
beanplot package
> crime <- read.csv("http://datasets.flowingdata.com/crimeRatesByState-formatted.csv")
> crime.new <- crime[crime$state != "District of Columbia",]
library(beanplot)
beanplot(crime.new[,-1],col="purple", ylab="prepared by VOLKAN OBAN" )

beanplot package
crime <- read.csv("http://datasets.flowingdata.com/crimeRatesByState-formatted.csv")
crime.new <- crime[crime$state != "District of Columbia",]
> library(beanplot)
> beanplot(crime.new[,-1])

tabplot in R.
library(tabplot)
> require(ggplot2)
> data(diamonds)
>
> tab <- tableplot(diamonds)
> plot(tab, title="prepared in R by VOLKAN OBAN \n data(diamonds)",
+ fontsize=12,
+ legend.lines=7,
+ fontsize.title=16)

Plot3D package-hist3D_fancy function
hist3D_fancy<- function(x, y, break.func = c("Sturges", "scott", "FD"), breaks = NULL,
colvar = NULL, col="white", clab=NULL, phi = 5, theta = 25, ...){
# Compute the number of classes for a histogram
break.func <- break.func [1]
if(is.null(breaks)){
x.breaks <- switch(break.func,
Sturges = nclass.Sturges(x),
scott = nclass.scott(x),
FD = nclass.FD(x))
y.breaks <- switch(break.func,
Sturges = nclass.Sturges(y),
scott = nclass.scott(y),
FD = nclass.FD(y))
} else x.breaks <- y.breaks <- breaks
# Cut x and y variables in bins for counting
x.bin <- seq(min(x), max(x), length.out = x.breaks)
y.bin <- seq(min(y), max(y), length.out = y.breaks)
xy <- table(cut(x, x.bin), cut(y, y.bin))
z <- xy
xmid <- 0.5*(x.bin[-1] + x.bin[-length(x.bin)])
ymid <- 0.5*(y.bin[-1] + y.bin[-length(y.bin)])
oldmar <- par("mar")
par (mar = par("mar") + c(0, 0, 0, 2))
hist3D(x = xmid, y = ymid, z = xy, ...,
zlim = c(-max(z)/2, max(z)), zlab = "counts", bty= "g",
phi = phi, theta = theta,
shade = 0.2, col = col, border = "black",
d = 1, ticktype = "detailed")
scatter3D(x, y,
z = rep(-max(z)/2, length.out = length(x)),
colvar = colvar, col = gg.col(100),
add = TRUE, pch = 18, clab = clab,
colkey = list(length = 0.5, width = 0.5,
dist = 0.05, cex.axis = 0.8, cex.clab = 0.8)
)
par(mar = oldmar)
}
data(iris)
hist3D_fancy(iris$Sepal.Length, iris$Petal.Width, main="prepared by Volkan OBAN ", colvar=as.numeric(iris$Species))

Plot3D package
set.seed(1234)
> x <- sort(rnorm(10))
> y <- runif(10)
> # Variable for coloring points
> col.v <- sqrt(x^2 + y^2)
> scatter2D(x, y, colvar = col.v, pch = 16, bty ="n",
+ type ="b")
> CI <- list()
> CI$x <- matrix(nrow = length(x), data = c(rep(0.25, 2*length(x))))
> scatter2D(x, y, colvar = col.v, pch = 16, bty ="n", cex = 1.5,
+ CI = CI, type = "b")
> CI <- list()
> CI$x <- matrix(nrow = length(x), data = c(rep(0.25, 2*length(x))))
> scatter2D(x, y, colvar = col.v, pch = 16, bty ="n", cex = 1.5, main="prepared in R bu Volkan OBAN - scatter2D function",
+ CI = CI, type = "b")

Plot3D package
ibrary(plot3D)
Warning message:
In as.list(X) : reached elapsed time limit
>
> X <- seq(0, 2*pi, length.out = 50)
> Y <- seq(-15, 6, length.out = 50)
> M <- mesh(X, Y)
> u <- M$x
> v <- M$y
>
> # x, y and z grids
> x <- (1.16 ^ v) * cos(v) * (1 + cos(u))
> y <- (-1.16 ^ v) * sin(v) * (1 + cos(u))
> z <- (-2 * 1.16 ^ v) * (1 + sin(u))
>
> # full colored image
> par(mai = c(0.01, 0.01, 0.01, 0.01))
> surf3D(x, y, z, colvar = z,
+ col = ramp.col(col = c("violet", "pink"), n = 100),
+ colkey = FALSE, shade = 0.5, alpha = 0.3, expand = 1.2,
+ box = FALSE, phi = 35, border = "black", theta = 70,
+ lighting = TRUE, ltheta = 560, lphi = -50)

R dsts viz
layout(matrix(c(1,1,1,2,3,4),nrow=2,ncol=3,byrow=TRUE))
# plot a proximity.timeline illustrating infection spread
proximity.timeline(toy_epi_sim,vertex.col = 'ndtvcol',
spline.style='color.attribute',
mode = 'sammon',default.dist=100,
chain.direction='reverse')
# plot 3 static cross-sectional networks
# (beginning, middle and end) underneath for comparison
plot(network.collapse(toy_epi_sim,at=1),vertex.col='ndtvcol',
vertex.cex=2,main='toy_epi_sim network at t=1')
plot(network.collapse(toy_epi_sim,at=17),vertex.col='ndtvcol',
vertex.cex=2,main='toy_epi_sim network at=17')
plot(network.collapse(toy_epi_sim,at=25),vertex.col='ndtvcol',
vertex.cex=2,main='toy_epi_sim network at t=25')
layout(1)
happy new year
> library(animation)
> library(picante)
> library(nlme)
> library(FD)
> library(vegan)
> library(permute)
> library(geometry)
> library(magic)
> library(abind)
> library(ape)
> library(ade4)
> fire <- function(centre = c(0, 0), r = 1:5, theta = seq(0,
+ 2 * pi, length = 100), l.col = rgb(1, 1, 0), lwd = 5,
+ ...) {
+ x <- centre[1] + outer(r, theta, function(r, theta) r *
+ sin(theta))
+ y <- centre[2] + outer(r, theta, function(r, theta) r *
+ cos(theta))
+ matplot(x, y, type = "l", lty = 1, col = l.col, add = T,
+ lwd = lwd, ...)
+ }
> f <- function(centre = rbind(c(-7, 7), c(7, 6)), n = c(7,
+ 5), N = 20, l.col = c("rainbow", "green"), p.col = "red",
+ lwd = 5, ...) {
+ ani.options(interval = 0.1)
+ lwd = lwd
+ if (is.vector(centre) && length(n) == 1) {
+ r = 1:n
+ l = seq(0.1, 0.6, length = n)
+ matplot(centre[1], centre[2], col = p.col, ...)
+ for (r in r) {
+ fire(centre = centre, r = seq(r - l[r], r + l[r],
+ length = 10), theta = seq(0, 2 * pi, length = 10 *
+ r) + 1, l.col = rainbow(n)[r], lwd = lwd, ...)
+ }
+ }
+ else {
+ matplot(centre[, 1], centre[, 2], col = p.col, ...)
+ l = list()
+ for (i in 1:length(n)) l[i] = list(seq(0.1, 0.6,
+ length = n[i]))
+ if (length(l.col) == 1)
+ l.col = rep(l.col, length(n))
+ r = 1:N
+ for (r in r) {
+ for (j in 1:length(n)) {
+ if (r%%(n[j] + 1) == 0) {
+ r1 = 1:n[j]
+ l1 = seq(0.1, 0.6, length = n[j])
+ for (r1 in r1) {
+ fire(centre = centre[j, ], r = seq(r1 -
+ l1[r1], r1 + l1[r1], length = 10), theta = seq(0,
+ 2 * pi, length = 10 * r1) + 1, l.col = par("bg"),
+ lwd = lwd + 2)
+ }
+ }
+ else {
+ if (l.col[j] == "red")
+ fire(centre = centre[j, ], r = seq(r%%(n[j] +
+ 1) - l[[j]][r%%(n[j] + 1)], r%%(n[j] +
+ 1) + l[[j]][r%%(n[j] + 1)], length = 10),
+ theta = seq(0, 2 * pi, length = 10 *
+ r%%(n[j] + 1)) + 1, l.col = rgb(1,
+ r%%(n[j] + 1)/n[j], 0), lwd = lwd,
+ ...)
+ else if (l.col[j] == "green")
+ fire(centre = centre[j, ], r = seq(r%%(n[j] +
+ 1) - l[[j]][r%%(n[j] + 1)], r%%(n[j] +
+ 1) + l[[j]][r%%(n[j] + 1)], length = 10),
+ theta = seq(0, 2 * pi, length = 10 *
+ r%%(n[j] + 1)) + 1, l.col = rgb(1 -
+ r%%(n[j] + 1)/n[j], 1, 0), lwd = lwd,
+ ...)
+ else if (l.col[j] == "blue")
+ fire(centre = centre[j, ], r = seq(r%%(n[j] +
+ 1) - l[[j]][r%%(n[j] + 1)], r%%(n[j] +
+ 1) + l[[j]][r%%(n[j] + 1)], length = 10),
+ theta = seq(0, 2 * pi, length = 10 *
+ r%%(n[j] + 1)) + 1, l.col = rgb(r%%(n[j] +
+ 1)/n[j], 0, 1), lwd = lwd, ...)
+ else fire(centre = centre[j, ], r = seq(r%%(n[j] +
+ 1) - l[[j]][r%%(n[j] + 1)], r%%(n[j] +
+ 1) + l[[j]][r%%(n[j] + 1)], length = 10),
+ theta = seq(0, 2 * pi, length = 10 * r%%(n[j] +
+ 1)) + 1, l.col = rainbow(n[j])[r%%(n[j] +
+ 1)], lwd = lwd, ...)
+ }
+ ani.pause()
+ }
+ }
+ }
+ }
> card <- function(N = 20, p.col = "green", bgcolour = "black",
+ lwd = 5, ...) {
+ ani.options(interval = 1)
+ for (i in 1:N) {
+ par(ann = F, bg = bgcolour, mar = rep(0, 4), pty = "s")
+ f(N = i, lwd = lwd, ...)
+ text(0, 0, "Happy New Year \n Happy New Year \n VOLKAN OBAN ", srt = 360 * i/N, col = rainbow(N)[i],
+ cex = 4.5 * i/N)
+ ani.pause()
+ }
+ }
> ani.options(interval = 0.2)
> card(N = 30, centre = rbind(c(-8, 8), c(8, 10), c(5, 0)), n = c(9, 5, 6), pch = 8, p.col = "green", l.col = c("rainbow", "red", "green"), xlim = c(-12, 12), ylim = c(-12,12))

wordcloud2 package
Turkish flag.Türk Bayrağı.
wordcloud2(demoFreq, figPath = "bayr.png", size = 1.5, color = "red", backgroundColor="white")

wordcloud2
ATATÜRK

word cloud2
wordcloud2(demoFreq, figPath = "ata.png", size = 1.5, color = "black", backgroundColor="white")

wordcloud2
Atatürk

wordcloud2 example .pi number
wordcloud2(demoFreq, figPath = "pii.png", size = 1.5, color = "skyblue", backgroundColor="black")

wordcloud2
wordcloud2(demoFreq, figPath = "atam.png", size = 1.5, color = "skyblue", backgroundColor="black")

wordcloud2
> library(wordcloud2)
> letterCloud( demoFreq, word = "itü", color='random-light' , backgroundColor="black")

wordcloud
library(wordcloud)
wordcloud(c("HAPPY NEW YEAR", "2017","VOLKAN OBAN"), max.words =100,min.freq=3,scale=c(4,.5), random.order = FALSE,rot.per=.5,vfont=c("gothic english","plain"),colors=palette())

wordcloud2
library(wordcloud2)
> letterCloud(demoFreq, word = "HAPPY NEW YEAR - 2017 !", wordSize = 1)

library(wordcloud2)
library(wordcloud2)
letterCloud( demoFreq, word = "2017 \ V. O. ", color='random-light' , backgroundColor="black")

ggparallel package
titanic data set.
Plot
library(ggplot2)
> #create data
> set.seed(3)
>
> #time steps
> t.step<-seq(0,20)
>
> #group names
> grps<-letters[1:10]
>
> #random data for group values across time
> grp.dat<-runif(length(t.step)*length(grps),5,15)
>
> #create data frame for use with plot
> grp.dat<-matrix(grp.dat,nrow=length(t.step),ncol=length(grps))
> grp.dat<-data.frame(grp.dat,row.names=t.step)
> names(grp.dat)<-grps
> source("https://gist.github.com/fawda123/6589541/raw/8de8b1f26c7904ad5b32d56ce0902e1d93b89420/plot_area.r")
>
> plot.area(grp.dat)
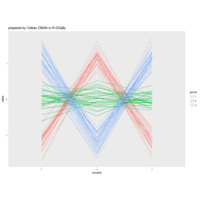
GGally
> set.seed(3674)
> k <- rep(1:3, each=30)
> x <- k + rnorm(mean=10, sd=.2,n=90)
> y <- -2*k + rnorm(mean=10, sd=.4,n=90)
> z <- 3*k + rnorm(mean=10, sd=.6,n=90)
>
> dat <- data.frame(group=factor(k),x,y,z)
>
> library(GGally)
> ggparcoord(dat,columns=1:4,groupColumn = 1)

ggplot2 example
> library(ggplot2)
> ggplot(data = diamonds) +
+ geom_bar(mapping = aes(x = cut, fill = cut), width = 1) +
+ coord_polar() +
+ facet_wrap( ~ clarity) + ggtitle("prepared in R by Volkan OBAN")
> ggplot(data = diamonds) +
+ geom_bar(mapping = aes(x = cut, fill = cut), width = 1) +
+ coord_polar() +
+ facet_wrap( ~ clarity) + ggtitle("prepared in R by Volkan OBAN \n data(diamonds)")

rastervVis package.
library(rasterVis)
> alt <- getData('worldclim', var='alt', res=2.5)
> a1 <- getData('GADM', country='Turkey', level=1)
> oregon <- a1[a1$NAME_1 == 'Oregon',]
> alt <- crop(alt, extent(oregon) + 0.5)
> alt <- mask(alt, oregon)
> levelplot(alt,main="prepared in R-rastervis package by Volkan OBAN \n TURKEY", par.settings=GrTheme)
magic Square 5*5
> m <- matrix(c(25,16,12,8,4,13,9,5,21,17,1,22,18,14,10,19,15,6,2,23,7,3,24,20,11), nrow=5, ncol=5)
> df <- expand.grid(x=1:ncol(m),y=1:nrow(m))
> df$val <- m[as.matrix(df[c('y','x')])]
> library(plotrix)
> xt <- xtabs(val ~ ., df[c(2,1,3)])
> color2D.matplot(xt, vcex = 3, show.values = 1, axes = FALSE, xlab = "Magic Square 5x5 ", ylab = "", cellcolors = rep("pink", length(xt)))
magic Square 5*5
Code:
m <- matrix(c(25,13,1,19,7,16,9,22,15,3,12,5,18,6,24,8,21,14,2,20,4,17,10,23,11), nrow=5, ncol=5)
> df <- expand.grid(x = 1:ncol(m),y = 1:nrow(m))
> df$val <- m[as.matrix(df[c('y', 'x')])]
> library(plotrix)
> xt <- xtabs(val ~ ., df[c(2,1,3)])
> color2D.matplot(xt, vcex = 3, show.values = 1, axes = FALSE, xlab = "Magic Square 5x5 ",
+ ylab = "prepared by Volkan OBAN ", cellcolors = rep("white", length(xt)))

Plotting
> x <- 1:400
> y <- sin(x/10) * exp(x * -0.01)
>
> plot(x, y)
> x <- 1:400
> y <- sin(x/10) * exp(x * -0.01)
>
> plot(x, y)
streamgraph in R.
data <- read.csv("http://bl.ocks.org/WillTurman/raw/4631136/data.csv", stringsAsFactors=FALSE)
data$date <- as.Date(data$date, format="%m/%d/%y")
streamgraph(data, interactive=TRUE) %>% sg_colors("Reds")
dat <- read.csv("http://asbcllc.com/blog/2015/february/cre_stream_graph_test/data/cre_transaction-data.csv")
dat %>%
streamgraph("asset_class", "volume_billions", "year", interpolate="cardinal") %>%
sg_axis_x(1, "year", "%Y") %>%
sg_fill_brewer("PuOr")
datatable(dat)
dat %>%
streamgraph("asset_class", "volume_billions", "year", offset="silhouette", interpolate="step") %>%
sg_axis_x(1, "year", "%Y") %>%
sg_fill_brewer("PuOr")
dat %>%
streamgraph("asset_class", "volume_billions", "year", offset="zero", interpolate="cardinal") %>%
sg_axis_x(1, "year", "%Y") %>%
sg_fill_brewer("PuOr") %>%
sg_legend(TRUE, "Asset class: ")
Now, who let that stacked bar chart get in here ;-)
dat %>%
streamgraph("asset_class", "volume_billions", "year", offset="zero", interpolate="step") %>%
sg_axis_x(1, "year", "%Y") %>%
sg_fill_brewer("PuOr")
# get top 10 names for each year by sex
babynames %>%
group_by(year, sex) %>%
top_n(10, n) -> dat1
# just look at female names and get the data for
# the top n by all years to see how they "flow"
babynames %>%
filter(sex=="F",
name %in% dat1$name) -> dat
streamgraph(dat, "name", "n", "year") %>%
sg_fill_tableau() %>%
sg_axis_x(tick_units = "year", tick_interval = 10, tick_format = "%Y") %>%
sg_legend(TRUE, "Name: ")

rbokeh example
> library(rbokeh)
> library(maps)
> data(world.cities)
> caps <- subset(world.cities, capital == 1)
> caps$population <- prettyNum(caps$pop, big.mark = ",")
> figure(width = 800, height = 450,title = "prepared by Volkan OBAN- rbokeh in R", padding_factor = 0) %>%
+ ly_map("world", col = "gray") %>%
+ ly_points(long, lat, data = caps, size = 5,
+ hover = c(name, country.etc, population))

Faceted Heatmap in R.
https://rpubs.com/omicsdata/faceted_heatmap

googleVis package. Calendar charts with googleVis
stock <- "MSFT"
start.date <- "2012-01-01"
end.date <- Sys.Date()
quote <- paste("http://ichart.finance.yahoo.com/table.csv?s=",
stock,
"&a=", substr(start.date,6,7),
"&b=", substr(start.date, 9, 10),
"&c=", substr(start.date, 1,4),
"&d=", substr(end.date,6,7),
"&e=", substr(end.date, 9, 10),
"&f=", substr(end.date, 1,4),
"&g=d&ignore=.csv", sep="")
stock.data <- read.csv(quote, as.is=TRUE)
stock.data$Date <- as.Date(stock.data$Date)
## Uncomment the next 3 lines to install the developer version of googleVis
# install.packages(c("devtools","RJSONIO", "knitr", "shiny", "httpuv"))
# library(devtools)
# install_github("mages/googleVis")
library(googleVis)
plot(
gvisCalendar(data=stock.data, datevar="Date", numvar="Adj.Close",
options=list(
title="Calendar heat map of MSFT adjsuted close",
calendar="{cellSize:10,
yearLabel:{fontSize:20, color:'#444444'},
focusedCellColor:{stroke:'red'}}",
width=590, height=320),
chartid="Calendar")
)
library(lattice)
> library(chron)
> source("http://blog.revolutionanalytics.com/downloads/calendarHeat.R")
> # Plot as calendar heatmap
> calendarHeat(stock.data$Date, stock.data$Adj.Close,
+ varname="PREPARED BY VOLKAN OBAN \n MSFT Adjusted Close")
> library(lattice)
> library(chron)
> source("http://blog.revolutionanalytics.com/downloads/calendarHeat.R")
> # Plot as calendar heatmap
> calendarHeat(stock.data$Date, stock.data$Adj.Close,
+ varname="\n PREPARED BY VOLKAN OBAN \n MSFT Adjusted Close")
> library(lattice)
> library(chron)
> source("http://blog.revolutionanalytics.com/downloads/calendarHeat.R")
> # Plot as calendar heatmap
> calendarHeat(stock.data$Date, stock.data$Adj.Close,
+ varname="MSFT Adjusted Close \n PREPARED BY VOLKAN OBAN \n")
>
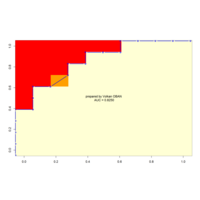
pROC package--Calculating AUC: the area under a ROC Curve
ref:https://www.r-bloggers.com/calculating-auc-the-area-under-a-roc-curve/

pROC package--Calculating AUC: the area under a ROC Curve
category <- c(1, 1, 1, 1, 0, 1, 1, 0, 1, 0, 1, 0, 1, 0, 0, 1, 0, 0, 0, 0)
prediction <- rev(seq_along(category))
prediction[9:10] <- mean(prediction[9:10])
library(pROC)
roc_obj <- roc(category, prediction)
auc(roc_obj)
## Area under the curve: 0.825
roc_df <- data.frame(
TPR=rev(roc_obj$sensitivities),
FPR=rev(1 - roc_obj$specificities),
labels=roc_obj$response,
scores=roc_obj$predictor)
rectangle <- function(x, y, width, height, density=12, angle=-45, ...)
polygon(c(x,x,x+width,x+width), c(y,y+height,y+height,y),
density=density, angle=angle, ...)
roc_df <- transform(roc_df,
dFPR = c(diff(FPR), 0),
dTPR = c(diff(TPR), 0))
plot(0:10/10, 0:10/10, type='n', xlab="FPR", ylab="TPR")
abline(h=0:10/10, col="lightblue")
abline(v=0:10/10, col="lightblue")
with(roc_df, {
mapply(rectangle, x=FPR, y=0,
width=dFPR, height=TPR, col="green", lwd=2)
mapply(rectangle, x=FPR, y=TPR,
width=dFPR, height=dTPR, col="blue", lwd=2)
lines(FPR, TPR, type='b', lwd=3, col="red")
})
ref:https://www.r-bloggers.com/calculating-auc-the-area-under-a-roc-curve/

lattice package-dotplot
library(lattice)
dotplot(VADeaths,groups=FALSE,layout=c(1,4),aspect=0.7,origin=0,type=c("p","h"),main="prepared by Volkan OBAN \n dotplot/Lattice package \n Death Rates in Virginia-1940",xlab="Rate (per 100)" )
Plots a phylogeny against the geological time scale-strap and geoscalePhylo package.
ref:https://rdrr.io/cran/strap/man/geoscalePhylo.html

Plots a phylogeny against the geological time scale-strap and geoscalePhylo package.
DatePhylo
geoscalePhylo

ggmap-Istanbul Technical University-my work place.
get_map
ggmap functions

flock-ggmap package in R.
Tur.map = get_map(location = "Turkey", zoom = 5, color="bw") ## get MAP data
p <- ggmap(Tur.map)
> p

plotly
> library(plotly)
>
> x <- c('Produce<br>Revenue', 'Services<br>Rev.', 'Total<br>Revenue', 'Fixed<br>Costs', 'Variable<br>Costs', 'Total<br>Costs', 'Total')
> y <- c(400, 660, 660, 590, 400, 400, 340)
> base <- c(0, 430, 0, 570, 370, 370, 0)
> revenue <- c(430, 260, 690, 0, 0, 0, 0)
> costs <- c(0, 0, 0, 120, 200, 320, 0)
> profit <- c(0, 0, 0, 0, 0, 0, 370)
> text <- c('$430K', '$260K', '$690K ', '$-1 20K', '$-200K', '$-320K', '$370K')
> data <- data.frame(x, base, revenue, costs, profit, text)
>
> #The default order will be alphabetized unless specified as below:
> data$x <- factor(data$x, levels = data[["x"]])
>
> p <- plot_ly(data, x = ~x, y = ~base, type = 'bar', marker = list(color = 'rgba(1,1,1, 0.0)')) %>%
+ add_trace(y = ~revenue, marker = list(color = 'rgba(55, 128, 191, 0.7)',
+ line = list(color = 'rgba(55, 128, 191, 0.7)',
+ width = 2))) %>%
+ add_trace(y = ~costs, marker = list(color = 'rgba(219, 64, 82, 0.7)',
+ line = list(color = 'rgba(219, 64, 82, 1.0)',
+ width = 2))) %>%
+ add_trace(y = ~profit, marker = list(color = 'rgba(50, 171, 96, 0.7)',
+ line = list(color = 'rgba(50, 171, 96, 1.0)',
+ width = 2))) %>%
+ layout(title = 'Annual Profit ',
+ xaxis = list(title = ""),
+ yaxis = list(title = "prepared by Volkan OBAN"),
+ barmode = 'stack',
+ paper_bgcolor = 'rgba(245, 246, 249, 1)',
+ plot_bgcolor = 'rgba(245, 246, 249, 1)',
+ showlegend = FALSE) %>%
+ add_annotations(text = text,
+ x = x,
+ y = y,
+ xref = "x",
+ yref = "y",
+ font = list(family = 'Arial',
+ size = 14,
+ color = 'rgba(245, 246, 249, 1)'),
+

sigma package in R
> library(devtools)
> devtools::install_github("jjallaire/sigma")
library(sigma)
> data <- system.file("examples/ediaspora.gexf.xml", package = "sigma")
> sigma(data)

rChartsCalendar package in R.
library(devtools)
install_github("ramnathv/rChartsCalendar")
dat <- read.csv('http://t.co/mN2RgcyQFc')[,c('date', 'pts')]
library(rChartsCalendar)
r1 <- plotCalMap(x = 'date', y = 'pts',
data = dat,
domain = 'month',
start = "2012-10-27",
legend = seq(10, 50, 10),
itemName = 'point',
range = 7
)
library(quantmod)
getSymbols("AAPL")
xts_to_df <- function(xt){
data.frame(
date = format(as.Date(index(xt)), '%Y-%m-%d'),
coredata(xt)
)
}
dat = xts_to_df(AAPL)
plotCalMap('date', 'AAPL.Adjusted',
data = dat,
domain = 'month',
legend = seq(500, 700, 40),
start = '2014-01-01',
itemName = '$$'
)
TimeProjection package in R.
library(TimeProjection)
>dates = timeSequence(from = '2012-01-01', to = '2012-12-31', by = 'day')
> plotCalendarHeatmap(as.Date(dates), 1:366

calendar heatmap.
stock.dailychange<-100*(diff(stock.data$Adj.Close,lag=1)/y[1:length(stock.data$Adj.Close)-1])
calendarHeat(stock.data$Date[1:length(stock.data$Date)-1], stock.dailychange, varname="SPY daily % changes(CL-CL)")

calendar heatmap.
code source: https://github.com/iascchen/VisHealth/blob/master/R/calendarHeat.R
then
stock <- "MSFT"
start.date <- "2006-01-12"
end.date <- Sys.Date()
quote <- paste("http://ichart.finance.yahoo.com/table.csv?s=",
stock,
"&a=", substr(start.date,6,7),
"&b=", substr(start.date, 9, 10),
"&c=", substr(start.date, 1,4),
"&d=", substr(end.date,6,7),
"&e=", substr(end.date, 9, 10),
"&f=", substr(end.date, 1,4),
"&g=d&ignore=.csv", sep="")
stock.data <- read.csv(quote, as.is=TRUE)
calendarHeat(stock.data$Date, stock.data$Adj.Close, varname="MSFT Adjusted Close")
qgraph package in R-an example.
library(qgraph)
dat.3 <- matrix(c(1:15*2-1,1:15*2),,2)
dat.3 <- cbind(dat.3,round(seq(-0.7,0.7,length=15),1))
# Create grid layout:
L.3 <- matrix(1:30,nrow=2)
# Different esize:
qgraph(dat.3,layout=L.3,directed=FALSE,edge.labels=TRUE,esize=14)
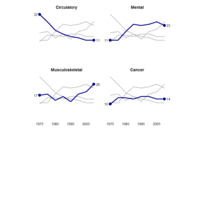
data visulazition in R an example
# split into 2 rows and 2 cols
split.screen(c(2,2))
# keep track of which screen we are
# plotting to
scr <- 1
# iterate over columns
for (i in 1:ncol(df)) {
# select screen
screen(scr)
# reduce margins
par(mar=c(3,2,1,1))
# empty plot
plot(1:nrow(df), 1:nrow(df), pch="", xlab=NA,
ylab=NA, xaxt="n", yaxt="n", ylim=c(0,35),
bty="n")
# plot all data in grey
for (j in 1:ncol(df)) {
lines(1:nrow(df), df[,j],
col="grey", lwd=3)
}
# plot selected in blue
lines(1:nrow(df), df[,i], col="blue4", lwd=4)
# add blobs
points(c(1,nrow(df)), c(df[1,i], df[nrow(df),i]),
pch=16, cex=2, col="blue4")
# add numbers
mtext(df[1,i], side=2, at=df[1,i], las=2)
mtext(df[nrow(df),i], side=4, at=df[nrow(df),i],
las=2)
# add title
title(colnames(df)[i])
# add axes if we are one of
# the bottom two plots
if (scr >= 3) {
axis(side=1, at=1:nrow(df), tick=FALSE,
labels=rownames(df))
}
# next screen
scr <- scr + 1
}
# close multi-panel image
close.screen(all=TRUE)

PerformanceAnalytics package
> library(PerformanceAnalytics)
> library(psych)
> d <- msq[,80:84]
> chart.Boxplot(d, main = "", xlab="average personality rating (based on n=3896) \n prepared by Volkan OBAN", ylab="",element.color = "transparent", as.Tufte=TRUE)
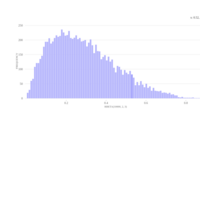
metricsgraphics
library(metricsgraphics)
library(RColorBrewer)
tmp <- data.frame(year=seq(1790, 1970, 10), uspop=as.numeric(uspop))
tmp %>%
mjs_plot(x=year, y=uspop) %>%
mjs_line() %>%
mjs_add_marker(1850, "Something Wonderful") %>%
mjs_add_baseline(150, "Something Awful")
tmp %>%
mjs_plot(x=year, y=uspop, width=600) %>%
mjs_line(area=TRUE)
tmp %>%
mjs_plot(x=uspop, y=year, width=500, height=400) %>%
mjs_bar() %>%
mjs_axis_x(xax_format = 'plain')
mtcars %>%
mjs_plot(x=wt, y=mpg, width=600, height=500) %>%
mjs_point(color_accessor=carb, size_accessor=carb) %>%
mjs_labs(x="Weight of Car", y="Miles per Gallon")
mtcars %>%
mjs_plot(x=wt, y=mpg, width=600, height=500) %>%
mjs_point(color_accessor=cyl,
x_rug=TRUE, y_rug=TRUE,
size_accessor=carb,
size_range=c(5, 10),
color_type="category",
color_range=brewer.pal(n=11, name="RdBu")[c(1, 5, 11)]) %>%
mjs_labs(x="Weight of Car", y="Miles per Gallon")
mtcars %>%
mjs_plot(x=wt, y=mpg, width=400, height=300) %>%
mjs_point(least_squares=TRUE) %>%
mjs_labs(x="Weight of Car", y="Miles per Gallon")
set.seed(1492)
dat <- data.frame(date=seq(as.Date("2014-01-01"),
as.Date("2014-01-31"),
by="1 day"),
value=rnorm(n=31, mean=0, sd=2))
dat %>%
mjs_plot(x=date, y=value) %>%
mjs_line() %>%
mjs_axis_x(xax_format = "date")
# Custom rollovers
dat %>%
mjs_plot(x=date, y=value) %>%
mjs_line() %>%
mjs_axis_x(xax_format = "date") %>%
mjs_add_mouseover("function(d, i) {
$('{{ID}} svg .mg-active-datapoint')
.text('custom text : ' + d.date + ' ' + i);
}")
# also works for scatterplots with a slight mod
set.seed(1492)
dat <- data.frame(value=rnorm(n=30, mean=5, sd=1),
value2=rnorm(n=30, mean=4, sd=1),
test = c(rep(c('test', 'test2'), 15)))
dat %>%
mjs_plot(x = value, y = value2) %>%
mjs_point() %>%
mjs_add_mouseover("function(d, i) {
$('{{ID}} svg .mg-active-datapoint')
.text('custom text : ' + d.point.test + ' ' + i);
}")
set.seed(1492)
stocks <- data.frame(
time = as.Date('2009-01-01') + 0:9,
X = rnorm(10, 0, 1),
Y = rnorm(10, 0, 2),
Z = rnorm(10, 0, 4))
stocks %>%
mjs_plot(x=time, y=X) %>%
mjs_line() %>%
mjs_axis_x(show=FALSE) %>%
mjs_axis_y(show=FALSE)
stocks %>%
mjs_plot(x=time, y=X) %>%
mjs_line() %>%
mjs_add_line(Y) %>%
mjs_add_line(Z) %>%
mjs_axis_x(xax_format="date")
mjs_plot(rnorm(10000)) %>%
mjs_histogram(bins=30, bar_margin=1)
movies <- ggplot2movies::movies[sample(nrow(ggplot2movies::movies), 1000), ]
mjs_plot(movies$rating) %>% mjs_histogram()
mjs_plot(movies, rating) %>%
mjs_histogram() %>%
mjs_labs(x_label="Histogram of movie ratings",
y_label="Frequency")
mjs_plot(movies$rating) %>% mjs_histogram(bins=30)
mjs_plot(runif(10000)) %>%
mjs_labs(x_label="runif(10000)") %>%
mjs_histogram()
mjs_plot(rbeta(10000, 2, 5)) %>%
mjs_labs(x_label="rbeta(10000, 2, 3)") %>%
mjs_histogram(bins=100) %>%
mjs_axis_y(extended_ticks=TRUE)
bimod <- c(rnorm(1000, 0, 1), rnorm(1000, 3, 1))
mjs_plot(bimod) %>% mjs_histogram()
mjs_plot(bimod) %>% mjs_histogram(bins=30)
bimod %>% mjs_hist(30)
library(shiny)
library(metricsgraphics)
ui = shinyUI(fluidPage(
h3("MetricsGraphics Example", style="text-align:center"),
metricsgraphicsOutput('mjs1'),
br(),
metricsgraphicsOutput('mjs2')
))
server = function(input, output) {
mtcars %>%
mjs_plot(x=wt, y=mpg, width=400, height=300) %>%
mjs_point(color_accessor=carb, size_accessor=carb) %>%
mjs_labs(x="Weight of Car", y="Miles per Gallon") -> m1
set.seed(1492)
stocks <- data.frame(
time = as.Date('2009-01-01') + 0:9,
X = rnorm(10, 0, 1),
Y = rnorm(10, 0, 2),
Z = rnorm(10, 0, 4))
stocks %>%
mjs_plot(x=time, y=X) %>%
mjs_line() %>%
mjs_add_line(Y) %>%
mjs_add_line(Z) %>%
mjs_axis_x(xax_format="date") %>%
mjs_add_legend(legend=c("X", "Y", "Z")) -> m2
output$mjs1 <- renderMetricsgraphics(m1)
output$mjs2 <- renderMetricsgraphics(m2)
}
shinyApp(ui = ui, server = server)

maps-geosphere in R
library(maps) # Provides functions that let us plot the maps
)
library('geosphere')
library(mapdata)
map('worldHires')
map("worldHires","Turkey",col="white", border="gray10", fill=TRUE, bg="paleturquoise1")

ggmap and mapproj
> library(ggmap)
> library(mapproj)
> map <- get_map(location = 'Europe', zoom = 4)
> ggmap(map)
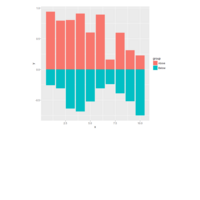
data visulazition in R
df <- data.frame(group = rep(c("Above", "Below"), each=10), x = rep(1:10, 2), y = c(runif(10, 0, 1), runif(10, -1, 0)))
> p <- ggplot(df, aes(x=x, y=y, fill=group)) +
+ geom_bar(stat="identity", position="identity")
> print(p)

library(PerformanceAnalytics)
library(PerformanceAnalytics)
chart.Correlation(iris[-5], bg=iris$Species, pch=21)
leaflet package in R.
3
4
5
6
7
8
9
10
11
12
#Library
library(leaflet)
# Background 1: NASA
m=leaflet() %>% addTiles() %>% setView( lng = 2.34, lat = 48.85, zoom = 5 ) %>% addProviderTiles("NASAGIBS.ViirsEarthAtNight2012")
m
# Background 2: World Imagery
m=leaflet() %>% addTiles() %>% setView( lng = 2.34, lat = 48.85, zoom = 3 ) %>% addProviderTiles("Esri.WorldImagery")
m

PLOT WITH AN IMAGE AS BACKGROUND--jpeg and ggplot2 .
Anadolu-Anatolia.
PLOT WITH AN IMAGE AS BACKGROUND--jpeg and ggplot2 .
> library(jpeg)
> my_image=readJPEG("itu.jpg")
> library(ggplot2)
Attaching package: ‘ggplot2’
The following object is masked _by_ ‘.GlobalEnv’:
midwest
> # Set up a plot area with no plot
> plot(1:2, type='n', main="", xlab="x", ylab="y")
>
> # Get the plot information so the image will fill the plot box, and draw it
> lim <- par()
> rasterImage(my_image, lim$usr[1], lim$usr[3], lim$usr[2], lim$usr[4])
> grid()
>
> #Add your plot !
> lines(c(1, 1.2, 1.4, 1.6, 1.8, 2.0), c(1, 1.3, 1.7, 1.6, 1.7, 1.0), type="b", lwd=5, col="white")
> # Set up a plot area with no plot
> plot(1:2, type='n', main="prepared by Volkan OBAN", xlab="x", ylab="y")
>
> # Get the plot information so the image will fill the plot box, and draw it
> lim <- par()
> rasterImage(my_image, lim$usr[1], lim$usr[3], lim$usr[2], lim$usr[4])
> grid()
>
> #Add your plot !
> lines(c(1, 1.2, 1.4, 1.6, 1.8, 2.0), c(1, 1.3, 1.7, 1.6, 1.7, 1.0), type="b", lwd=5, col="black")

plotwidgetGallery
> plotwidgetGallery()
> ## automatically set black bg
> plotwidgetGallery(theme="neon")
> ## yuck, ugly:
> plotwidgetGallery(pal=c("red", "#FF9900", "blue", "green", "cyan", "yellow"))
> ## much better:
> plotwidgetGallery(pal=plotPals("pastel", alpha=0.8))

plotwidgets
plot.new()
## Loop over a few saturation / lightess values
par(usr=c(-0.5, 0.5, -0.5, 0.5))
v <- c(10, 9, 19, 9, 15, 5)
pal <- plotPals("zeileis")
for(sat in seq.int(-0.4, 0.4, length.out=5)) {
for(lgh in seq.int(-0.4, 0.4, length.out=5)) {
cols <- saturateCol(darkenCol(pal, by=sat), by=lgh)
wgPlanets(x=sat, y=lgh, w=0.16, h=0.16, v=v, col=cols)
}
}
axis(1)
axis(2)
title(xlab="Darkness (L) by=", ylab="Saturation (S) by=")
## Now loop over hues
a2xy <- function(a, r=1, full=FALSE) {
t <- pi/2 - 2 * pi * a / 360
list( x=r * cos(t), y=r * sin(t) )
}
plot.new()
par(usr=c(-1,1,-1,1))
hues <- seq(0, 360, by=30)
pos <- a2xy(hues, r=0.75)
for(i in 1:length(hues)) {
cols <- modhueCol(pal, by=hues[i])
wgPlanets(x=pos$x[i], y=pos$y[i], w=0.5, h=0.5, v=v, col=cols)
}
pos <- a2xy(hues[-1], r=0.4)
text(pos$x, pos$y, hues[-1])
rpivotTable package in R.
> rpivotTable(
+ Titanic,
+ rows = "Survived",
+ cols = c("Class","Sex"),
+ aggregatorName = "Sum as Fraction of Columns",
+ vals = "Freq",
+ rendererName = "Table Barchart"
+ )
>
> # An example with inclusions and exclusions filters:
>
> rpivotTable(
+ Titanic,
+ rows = "Survived",
+ cols = c("Class","Sex"),
+ aggregatorName = "Sum as Fraction of Columns",
+ inclusions = list( Survived = list("Yes")),
+ exclusions= list( Class = list( "Crew")),
+ vals = "Freq",
+ rendererName = "Table Barchart"
+ )
rpivotTable package in R.
daata(Titanic)
rpivotTable package in R.
> library(rpivotTable)
> data(mtcars)
> ## One line to create pivot table
> rpivotTable(mtcars, rows="gear", col="cyl", aggregatorName="Average", vals="mpg", rendererName="Treemap")
rpivotTable package in R.
> library(rpivotTable)
> data(mtcars)
> ## One line to create pivot table
> rpivotTable(mtcars, rows="gear", col="cyl", aggregatorName="Average", vals="mpg", rendererName="Treemap")

bwplot-lattice package.
> bwplot(~weight|factor(Time),data=ChickWeight,col="blue",
main="prepared by Volkan OBAN \n Weight by Days Since Birth",xlab="Weight in grams")

bwplot-lattice package.
library(lattice)
> bwplot(height~voice.part, singer)
> at <- seq(60, 75, 2.5)
> bwplot(height~voice.part, singer, scales=list(y=list(at=at)))
> bwplot(height~voice.part, singer, scales=list(y=list(at=at)),
+ panel=function(...) {
+ panel.abline(h=at, col="gray")
+ panel.bwplot(...)
+ })
> lvls <- levels(singer$voice.part)
> fill <- rep("white", length(lvls))
> fill[lvls %in% c("Tenor 1", "Tenor 2")] <- "gray"
> bwplot(height~voice.part, singer, scales=list(y=list(at=at)),
+ fill=fill, panel=function(...) {
+ panel.abline(h=at, col="gray")
+ panel.bwplot(...)
+ })

genhistogram-genasis package in R
library(genasis)
genhistogram(rnorm(60))
## Use of example data from the package:
data(kosetice.pas.openair)
genhistogram(kosetice.pas.openair[,1:8],col="orange",emboss=3)
data(kosetice.pas.genasis)
genhistogram(kosetice.pas.genasis[1:208,],input="genasis",
distr="lnorm",col="orange",emboss=2)

highcharter package.
highchart() %>%
+ hc_chart(backgroundColor = "#") %>%
+ hc_title(text = "Chart color gradient it's on fire-prepared by Volkan OBAN", style = list(color = "#CCC")) %>%
+ # hc_xAxis(categories = month.abb) %>%
+ hc_yAxis(labels = list(style = list(color = "#CCC")),
+ gridLineColor = "#111111") %>%
+ hc_series(
+ list(
+ data = abs(rnorm(100)) + 1,
+ type = "areaspline",
+ marker = list(enabled = FALSE),
+ color = list(
+ linearGradient = list(x1 = 0, y1 = 1, x2 = 0, y2 = 0),
+ stops = list(
+ list(0, "transparent"),
+ list(0.33, "yellow"),
+ list(0.66, "red"),
+ list(1, "#ccc")
+ )
+ ),
+ fillColor = list(
+ linearGradient = list(x1 = 0, y1 = 1, x2 = 0, y2 = 0),
+ stops = list(
+ list(0, "transparent"),
+ list(0.1, "yellow"),
+ list(0.5, "red"),
+ list(1, "black")
+ )
+ )
+ )
+ )
>
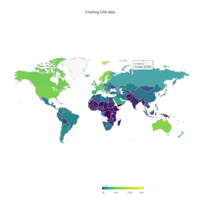
highcharter package.
data(worldgeojson)
data(GNI2014, package = "treemap")
dshmstops <- data.frame(q = c(0, exp(1:5)/exp(5)), c = substring(viridis(5 + 1), 0, 7)) %>%
list.parse2()
highchart() %>%
hc_title(text = "Charting GNI data") %>%
hc_add_series_map(worldgeojson, GNI2014,
value = "GNI", joinBy = "iso3") %>%
hc_colorAxis(stops = dshmstops)

highcharter package.
> highchart() %>%
+ hc_title(text = "prepared by Volkan OBAN-highcharter package") %>%
+ hc_xAxis(categories = month.abb) %>%
+ hc_defs(patterns = list(
+ list(id = 'custom-pattern',
+ path = list(d = 'M 0 0 L 10 10 M 9 -1 L 11 1 M -1 9 L 1 11',
+ stroke = "black",
+ strokeWidth = 1
+ )
+ )
+ )) %>%
+ hc_add_series(data = c(7.0, 6.9, 9.5, 14.5, 18.2, 21.5, 25.2,
+ 26.5, 23.3, 18.3, 13.9, 9.6),
+ type = "area",
+ fillColor = 'url(#custom-pattern)') %>%
+ hc_add_theme(hc_theme_handdrawn())
highcharter package.
library("MASS")
dscars <- round(mvrnorm(n = 20, mu = c(1, 1), Sigma = matrix(c(1,0,0,1),2)), 2)
dsplan <- round(mvrnorm(n = 10, mu = c(3, 4), Sigma = matrix(c(2,.5,2,2),2)), 2)
dstrck <- round(mvrnorm(n = 15, mu = c(5, 1), Sigma = matrix(c(1,.5,.5,1),2)), 2)
highchart() %>%
hc_chart(type = "scatter", zoomType = "xy") %>%
hc_tooltip(
useHTML = TRUE,
pointFormat = paste0("<span style=\"color:{series.color};\">{series.options.icon}</span>",
"{series.name}: <b>[{point.x}, {point.y}]</b><br/>")
) %>%
hc_add_series(data = list_parse2(as.data.frame(dscars)),
marker = list(symbol = fa_icon_mark("car")),
icon = fa_icon("car"), name = "car") %>%
hc_add_series(data = list_parse2(as.data.frame(dsplan)),
marker = list(symbol = fa_icon_mark("plane")),
icon = fa_icon("plane"), name = "plane") %>%
hc_add_series(data = list_parse2(as.data.frame(dstrck)),
marker = list(symbol = fa_icon_mark("truck")),
icon = fa_icon("truck"), name = "truck")

highcharter package.
SPY <- getSymbols("SPY", from="2015-01-01", auto.assign=FALSE)
SPY <- adjustOHLC(SPY)
SPY.SMA.10 <- SMA(Cl(SPY), n=10)
SPY.SMA.200 <- SMA(Cl(SPY), n=200)
SPY.RSI.14 <- RSI(Cl(SPY), n=14)
SPY.RSI.SellLevel <- xts(rep(70, NROW(SPY)), index(SPY))
SPY.RSI.BuyLevel <- xts(rep(30, NROW(SPY)), index(SPY))
highchart() %>%
# create axis :)
hc_yAxis_multiples(
list(title = list(text = NULL), height = "45%", top = "0%"),
list(title = list(text = NULL), height = "25%", top = "47.5%", opposite = TRUE),
list(title = list(text = NULL), height = "25%", top = "75%")
) %>%
# series :D
hc_add_series_ohlc(SPY, yAxis = 0, name = "SPY") %>%
hc_add_series_xts(SPY.SMA.10, yAxis = 0, name = "Fast MA") %>%
hc_add_series_xts(SPY.SMA.200, yAxis = 0, name = "Slow MA") %>%
hc_add_series_xts(SPY$SPY.Volume, color = "gray", yAxis = 1, name = "Volume", type = "column") %>%
hc_add_series_xts(SPY.RSI.14, yAxis = 2, name = "Osciallator") %>%
hc_add_series_xts(SPY.RSI.SellLevel, color = "red", yAxis = 2, name = "Sell level", enableMouseTracking = FALSE) %>%
hc_add_series_xts(SPY.RSI.BuyLevel, color = "blue", yAxis = 2, name = "Buy level", enableMouseTracking = FALSE) %>%
# I <3 themes
hc_add_theme(hc_theme_smpl())
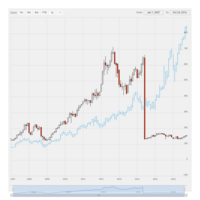
highcharter package.
SPY <- getSymbols("SPY", from="2015-01-01", auto.assign=FALSE)
SPY <- adjustOHLC(SPY)
SPY.SMA.10 <- SMA(Cl(SPY), n=10)
SPY.SMA.200 <- SMA(Cl(SPY), n=200)
SPY.RSI.14 <- RSI(Cl(SPY), n=14)
SPY.RSI.SellLevel <- xts(rep(70, NROW(SPY)), index(SPY))
SPY.RSI.BuyLevel <- xts(rep(30, NROW(SPY)), index(SPY))
highchart() %>%
# create axis :)
hc_yAxis_multiples(
list(title = list(text = NULL), height = "45%", top = "0%"),
list(title = list(text = NULL), height = "25%", top = "47.5%", opposite = TRUE),
list(title = list(text = NULL), height = "25%", top = "75%")
) %>%
# series :D
hc_add_series_ohlc(SPY, yAxis = 0, name = "SPY") %>%
hc_add_series_xts(SPY.SMA.10, yAxis = 0, name = "Fast MA") %>%
hc_add_series_xts(SPY.SMA.200, yAxis = 0, name = "Slow MA") %>%
hc_add_series_xts(SPY$SPY.Volume, color = "gray", yAxis = 1, name = "Volume", type = "column") %>%
hc_add_series_xts(SPY.RSI.14, yAxis = 2, name = "Osciallator") %>%
hc_add_series_xts(SPY.RSI.SellLevel, color = "red", yAxis = 2, name = "Sell level", enableMouseTracking = FALSE) %>%
hc_add_series_xts(SPY.RSI.BuyLevel, color = "blue", yAxis = 2, name = "Buy level", enableMouseTracking = FALSE) %>%
# I <3 themes
hc_add_theme(hc_theme_smpl())
highcharter package.-hcart
data(mpg)
library(dplyr)
mpgman3 <- group_by(mpg, manufacturer) %>%
+ summarise(n = n(), unique = length(unique(model))) %>%
+ arrange(-n, -unique)
hchart(mpgman3, "treemap", x = manufacturer, value = n, color = unique)

cartography package in R.
> nuts2.df$cagr <- (((nuts2.df$pop2008 / nuts2.df$pop1999)^(1/9)) - 1) * 100
> summary(nuts2.df$cagr)
Min. 1st Qu. Median Mean 3rd Qu. Max.
-2.42900 -0.08116 0.27750 0.31550 0.65960 3.02800
> # Plot the compound annual growth rate
> cols <- carto.pal(pal1 = "blue.pal", n1 = 2, pal2 = "red.pal", n2 = 4)
> choroLayer(spdf = nuts2.spdf,
+ df = nuts2.df,
+ var = "cagr", breaks = c(-2.43,-1,0,0.5,1,2,3.1),
+ col = cols,
+ border = "grey40",
+ add = FALSE,
+ legend.pos = "topright",legend.title.txt = "Compound annual\ngrowth rate",
+ legend.values.rnd = 2)
> # Layout plot
> layoutLayer(title = " CARTOGRAPHY package in R.prepared by Volkan OBAN \n Demographic Trends",
+ sources = "Eurostat, 2008",
+ scale = NULL,
+ frame = TRUE,
+ col = "black",
+ coltitle = "white")

lattice example
> dp.uspe <-
dotplot(t(USPersonalExpenditure), groups = FALSE, layout = c(1, 5),
xlab = "Expenditure (billion dollars)")
> dp.uspe.log <-
dotplot(t(USPersonalExpenditure), groups = FALSE, layout = c(1, 5),
scales = list(x = list(log = 2)),
xlab = "Expenditure (billion dollars)")
> plot(dp.uspe, split = c(1, 1, 2, 1))
> plot(dp.uspe.log, split = c(2, 1, 2, 1), newpage = FALSE)

lattice example
library(lattice)
> VADeaths
Rural Male Rural Female Urban Male Urban Female
50-54 11.7 8.7 15.4 8.4
55-59 18.1 11.7 24.3 13.6
60-64 26.9 20.3 37.0 19.3
65-69 41.0 30.9 54.6 35.1
70-74 66.0 54.3 71.1 50.0
> VADeathsDF <- as.data.frame.table(VADeaths, responseName = "Rate")
> VADeathsDF
Var1 Var2 Rate
1 50-54 Rural Male 11.7
2 55-59 Rural Male 18.1
3 60-64 Rural Male 26.9
4 65-69 Rural Male 41.0
5 70-74 Rural Male 66.0
6 50-54 Rural Female 8.7
7 55-59 Rural Female 11.7
8 60-64 Rural Female 20.3
9 65-69 Rural Female 30.9
10 70-74 Rural Female 54.3
11 50-54 Urban Male 15.4
12 55-59 Urban Male 24.3
13 60-64 Urban Male 37.0
14 65-69 Urban Male 54.6
15 70-74 Urban Male 71.1
16 50-54 Urban Female 8.4
17 55-59 Urban Female 13.6
18 60-64 Urban Female 19.3
19 65-69 Urban Female 35.1
20 70-74 Urban Female 50.0
> barchart(Var1 ~ Rate | Var2, VADeathsDF, layout = c(4, 1))
>

lattice
bwplot(gcsescore ~ gender | factor(score), Chem97, layout = c(6, 1))

lattice
> data(Chem97, package = "mlmRev")
> qqmath(~ gcsescore | factor(score), Chem97, groups = gender,
+ f.value = ppoints(100), auto.key = TRUE,
+ type = c("p", "g"), aspect = "xy")
>
> bwplot(factor(score) ~ gcsescore | gender, Chem97)
LatticeExtra
depth.ord <- rev(order(quakes$depth))
quakes$Magnitude <- equal.count(quakes$mag, 4)
quakes.ordered <- quakes[depth.ord, ]
levelplot(depth ~ long + lat | Magnitude, data = quakes.ordered,
panel = panel.levelplot.points, type = c("p", "g"),
aspect = "iso", prepanel = prepanel.default.xyplot)
## a levelplot with jittered cells
xyz <- expand.grid(x = 0:9, y = 0:9)
xyz[] <- jitter(as.matrix(xyz))
xyz$z <- with(xyz, sqrt((x - 5)^2 + (y - 5)^2))
levelplot(z ~ x * y, xyz, panel = panel.voronoi, points = FALSE)
## hexagonal cells
xyz$y <- xyz$y + c(0, 0.5)
levelplot(z ~ x * y, xyz, panel = panel.voronoi, points = FALSE)

lattice, grid, spatstat
# load required libraries
library(spatstat)
library(lattice)
library(grid)
library(CircStats)
# read in our data (see attached file)
x <- read.csv('beer_battle.csv')
# plot the data, as stratified by person
xyplot(y ~ x | person, groups=beer, data=x, panel=panel.bulls_eye,
key=list(points=list(col=c(1,2,3), pch=c(3,3,3)), text=list(c('0 beers', '1 beer', '3 beers')), columns=3),
main='Beer Battle 1'
)
gstat, LatticeExtra, grid package
library(gstat)
library(latticeExtra)
library(grid)
# load example data
data(meuse.grid)
data(meuse)
data(meuse.alt)
coordinates(meuse.grid) <- ~ x + y
coordinates(meuse) <- ~ x + y
coordinates(meuse.alt) <- ~ x + y
# converto SpatialPixelsDataFram
gridded(meuse.grid) <- TRUE
# convert 'soil' to factor and re-label
meuse.grid$soil <- factor(meuse.grid$soil, labels=c('A','B','C'))
meuse$soil <- factor(meuse$soil, levels=c('1','2','3'), labels=c('A','B','C'))
# setup color scheme
cols <- brewer.pal(n=3, 'Set1')
p.pch <- c(2,3,4)
# generate list of trellis settings
tps <- list(regions=list(col=cols), superpose.polygon=list(col=cols), superpose.symbol=list(col='black', pch=p.pch))
# init list of overlays
spl <- list('sp.points', meuse, cex=0.75, pch=p.pch[meuse$soil], col='black')
# setup trellis options
trellis.par.set(tps)
# initial plot, missing key
p1 <- spplot(meuse.grid, 'soil', sp.layout=spl, colorkey=FALSE, col.regions=cols, cuts=length(cols)-1)
# add a key at the top + space for key
p1 <- update(p1, key=simpleKey(levels(meuse.grid$soil), points=FALSE, lines=FALSE, rect=TRUE, regions=TRUE, columns=3, title='Class', cex=0.75))
# add a key on the right + space for key
p1 <- update(p1, key=simpleKey(levels(meuse$soil), points=TRUE, columns=1, title='Class', cex=0.75, space='right', ))
p1
.........
.....

ggplot2 example
> set.seed(654)
> week <- sample(0:9, 3000, rep=TRUE, prob = rchisq(10, df = 3))
> status <- factor(rbinom(3000, 1, 0.15), labels = c("Shipped", "Not-Shipped"))
> data.df <- data.frame(Week = week, Status = status)
> library("plyr")
> plot.df <- ddply(data.df, .(Week, Status), nrow)
> plot.df$V1 <- ifelse(plot.df$Status == "Shipped",
+ plot.df$V1, -plot.df$V1)
> library("ggplot2")
> ggplot(plot.df) +
+ aes(x = as.factor(Week), y = V1, fill = Status) +
+ geom_bar(stat = "identity", position = "identity") +
+ scale_y_continuous(breaks = 100 * -1:5,
+ labels = 100 * c(1, 0:5)) +
+ geom_text(aes(y = sign(V1) * max(V1) / 30, label = abs(V1)))
> r<-ggtitle("prepared by Volkan OBAN")
> library("ggplot2")
> ggplot(plot.df) +
+ aes(x = as.factor(Week), y = V1, fill = Status) +
+ geom_bar(stat = "identity", position = "identity") +
+ scale_y_continuous(breaks = 100 * -1:5,
+ labels = 100 * c(1, 0:5)) +
+ geom_text(aes(y = sign(V1) * max(V1) / 30, label = abs(V1)))

Pyramid plot in R
library(XML)
> library(reshape2)
> library(plyr)
> library(ggplot2)
> source('http://klein.uk/R/Viz/pyramids.R')
> popGHcens <- getAgeTable(country = "QA", year = 2015)
>
> pyramidGH <- ggplot(popGHcens, aes(x = Age, y = Population, fill = Gender)) +
+ geom_bar(data = subset(popGHcens, Gender == "Female"), stat = "identity") +
+ geom_bar(data = subset(popGHcens, Gender == "Male"), stat = "identity") +
+ scale_y_continuous(labels = paste0(as.character(c(seq(2, 0, -1), seq(1, 2, 1))), "m")) +
+ coord_flip()
> pyramidGH
ggplot2
> test <- (data.frame(v=rnorm(1000), g=c('M','F')))
> require(ggplot2)
> ggplot(data=test, aes(x=v)) +
+ geom_histogram() +
+ coord_flip() +
+ facet_grid(. ~ g)
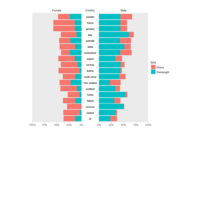
Pyramid plot in R
library(plotrix)
xy.males.overweight<-c(23.2,33.5,43.6,33.6,43.5,43.5,43.9,33.7,53.9,43.5,43.2,42.8,22.2,51.8,
41.5,31.3,60.7,50.4)
xx.females.overweight<-c(13.2,9.4,13.5,13.5,13.5,23.7,8,3.18,3.9,3.16,23.2,22.5,22,12.7,12.5,
12.3,10,0.8)
agelabels<-c("uk","scotland","france","ireland","germany","sweden","norway",
"iceland","portugal","austria","switzerland","australia","new zealand","dubai","south africa",
"finland","italy","morocco")
par(mar=pyramid.plot(xy.males.overweight,xx.females.overweight,labels=agelabels,
gap=9))
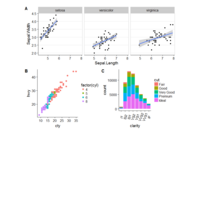
cowplot package--ggdraw
plot.iris <- ggplot(iris, aes(Sepal.Length, Sepal.Width)) +
geom_point() + facet_grid(. ~ Species) + stat_smooth(method = "lm") +
background_grid(major = 'y', minor = "none") + # add thin horizontal lines
panel_border() # and a border around each panel
# plot.mpg and plot.diamonds were defined earlier
ggdraw() +
draw_plot(plot.iris, 0, .5, 1, .5) +
draw_plot(plot.mpg, 0, 0, .5, .5) +
draw_plot(plot.diamonds, .5, 0, .5, .5) +
draw_plot_label(c("A", "B", "C"), c(0, 0, 0.5), c(1, 0.5, 0.5), size = 15)

ggdraw
boxes <- data.frame(
x = sample((0:36)/40, 40, replace = TRUE),
y = sample((0:32)/40, 40, replace = TRUE)
)
# plot on top of annotations
ggdraw() +
geom_rect(data = boxes, aes(xmin = x, xmax = x + .15, ymin = y, ymax = y + .15),
colour = "gray60", fill = "gray80") +
draw_plot(plot.mpg) +
draw_label("Plot is on top of the grey boxes", x = 1, y = 1,
vjust = 1, hjust = 1, size = 10, fontface = 'bold')
# plot below annotations
ggdraw(plot.mpg) +
geom_rect(data = boxes, aes(xmin = x, xmax = x + .15, ymin = y, ymax = y + .15),
colour = "gray60", fill = "gray80") +
draw_label("Plot is underneath the grey boxes", x = 1, y = 1,
vjust = 1, hjust = 1, size = 10, fontface = 'bold')

cowplot package.
> require(cowplot)
> plot.mpg <- ggplot(mpg, aes(x = cty, y = hwy, colour = factor(cyl))) +
+ geom_point(size=2.5)
>plot.diamonds <- ggplot(diamonds, aes(clarity, fill = cut)) + geom_bar() +
+ theme(axis.text.x = element_text(angle=70, vjust=0.5))
>plot_grid(plot.mpg, plot.diamonds, labels = c("A", "B"))

caret-R graphic.
> library(caret)
Zorunlu paket yükleniyor: lattice
> # load the iris dataset
> data(iris)
> x <- iris[,1:4]
> y <- iris[,5]
> featurePlot(x=x, y=y, plot="box")
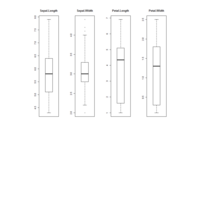
boxplot-ggplot2
> library(ggplot2)
> data(iris)
> par(mfrow=c(1,4))
> for(i in 1:4) {
boxplot(iris[,i], main=names(iris)[i])
}
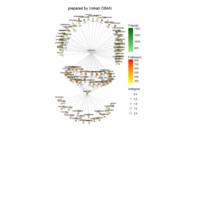
GGally-ggplot2
>library(GGally)
>data(twitter_spambots)
> ggnetworkmap(net = twitter_spambots,
+ arrow.size = 0.5,
+ node.group = followers,
+ ring.group = friends,
+ size = 4,
+ weight = indegree,
+ label.nodes = TRUE, vjust = -1.5) +
+ scale_fill_continuous("Followers", high = "red", low = "yellow") +
+ labs(color = "Friends") +
+ scale_color_continuous(low = "lightgreen", high = "darkgreen")
spie Chart--library(caroline)
library(caroline)

LatticeExtra-barchart
> library(latticeExtra)
> data(postdoc)
> library(lattice)
> barchart(prop.table(postdoc, margin = 1),
+ auto.key = TRUE, xlab = "Proportion")

stripchart
ggplot2
data(airquality)
> stripchart(Temp~Month,
+ data=airquality,
+ main="Different strip chart for each month",
+ xlab="Months",
+ ylab="Temperature",
+ col="brown3",
+ group.names=c("May","June","July","August","September"),
+ vertical=TRUE,
+ pch=16
+ )

nlme package in R.
> # There are 12 cities
> n.cities <- 12
>
> # The area of those cities (more reasonably, the logarithm
> # of their areas) are gaussian, independant variables.
> area.moyenne <- 5
> area.sd <- 1
> area <- rnorm(n.cities, area.moyenne, area.sd)
>
> a <- rnorm(n.cities)
> b <- rnorm(n.cities)
>
> # 200 inhabitants sampled in each city
> n.inhabitants <- 20
> city <- rep(1:n.cities, each=n.inhabitants)
>
> # The age are independant gaussian variables, mean=40, sd=10
> # We could have chosen a different distribution for each city.
> # (either randomly, or depending on their area or population).
>
> age <- rnorm(n.cities*n.inhabitants, 40, 10)
>
> # The income (the variable we try to explain) is a function of the
> # age, but the coefficients depend on the city
> # Here, the coefficients are taken at random, but they could
> # depend on the city area or population.
> # Here, the coefficients are independant -- this is rarely the case
> a <- rnorm(n.cities, 20000, sd=2000)
> b <- rnorm(n.cities, sd=20)
> income <- 200*area[city] + a[city] + b[city]*age +
+ rnorm(n.cities*n.inhabitants, sd=200)
>
> plot(income ~ age, col=rainbow(n.cities)[city], pch=16)
library(nlme)
d <- data.frame(income, age, city, area=area[city])
r <- lmList(income ~ age | city, data=d)
plot(intervals(r))
example-plot in R
# There are 12 cities
n.cities <- 12
# The area of those cities (more reasonably, the logarithm
# of their areas) are gaussian, independant variables.
area.moyenne <- 5
area.sd <- 1
area <- rnorm(n.cities, area.moyenne, area.sd)
a <- rnorm(n.cities)
b <- rnorm(n.cities)
# 200 inhabitants sampled in each city
n.inhabitants <- 20
city <- rep(1:n.cities, each=n.inhabitants)
# The age are independant gaussian variables, mean=40, sd=10
# We could have chosen a different distribution for each city.
# (either randomly, or depending on their area or population).
age <- rnorm(n.cities*n.inhabitants, 40, 10)
# The income (the variable we try to explain) is a function of the
# age, but the coefficients depend on the city
# Here, the coefficients are taken at random, but they could
# depend on the city area or population.
# Here, the coefficients are independant -- this is rarely the case
a <- rnorm(n.cities, 20000, sd=2000)
b <- rnorm(n.cities, sd=20)
income <- 200*area[city] + a[city] + b[city]*age +
rnorm(n.cities*n.inhabitants, sd=200)
plot(income ~ age, col=rainbow(n.cities)[city], pch=16)
Hierarchical anova --plot
> n <- 2000 # Number of experiments
> k <- 20 # Number of subjects
> l <- 4 # Number of groups
> kl <- sample(1:l, k, replace=T) # Group of each subject
> x1 <- sample(1:k, n, replace=T)
> x2 <- kl[x1]
> A <- rnorm(1,sd=4)
> B <- rnorm(k,sd=4)
> C <- rnorm(l,sd=4)
> y <- A + B[x1] + C[x2] + rnorm(n)
> x1 <- factor(x1)
> x2 <- factor(x2)
> op <- par(mfrow=c(1,2))
> plot(y~x1, col='pink')
> plot(y~x2, col='pink')
> par(op)
> mtext("Hierarchical anova", line=1.5, font=2, cex=1.2)
> # If the data were real, we wouldn't know kl.
> # One may recover it that way.
> kl <- tapply(x2,
+ x1,
+ function (x) {
+ a <- table(x)
+ names(a)[which(a==max(a))[1]]
+ })
> kl <- factor(kl, levels=levels(x2))
> plot( y ~ x1, col = rainbow(l)[kl],
+ main = "Hierarchical anova")
Hierarchical anova --plot
> n <- 2000 # Number of experiments
> k <- 20 # Number of subjects
> l <- 4 # Number of groups
> kl <- sample(1:l, k, replace=T) # Group of each subject
> x1 <- sample(1:k, n, replace=T)
> x2 <- kl[x1]
> A <- rnorm(1,sd=4)
> B <- rnorm(k,sd=4)
> C <- rnorm(l,sd=4)
> y <- A + B[x1] + C[x2] + rnorm(n)
> x1 <- factor(x1)
> x2 <- factor(x2)
> op <- par(mfrow=c(1,2))
> plot(y~x1, col='pink')
> plot(y~x2, col='pink')
> par(op)
> mtext("Hierarchical anova", line=1.5, font=2, cex=1.2)

Example.
> library(ggplot2)
>
> this_base <- "playfair-s-population-of-cities"
>
> my_data <- data.frame(
+ population = c(60, 63, 75, 80, 80, 80, 90, 120,
+ 130, 140, 145, 160, 180, 200, 210,
+ 220, 250, 255, 380, 690, 900, 1100),
+ city = c('Istanbul', 'Izmir', 'Erzurum', 'Kars', 'Manisa',
+ 'Muğla', 'Bursa', 'Antalya', 'Mersin', 'Rize',
+ 'Van', 'Adıyaman', 'Ankara', 'Kayseri', 'Trabzon',
+ 'Muş', 'Balıkesir', 'Sinop', 'Kastamonu', 'Aydın',
+ 'Isparta', 'İzmit'))
>
> # include helper variables
> my_data$row <- c(rep("row1", 11), rep("row2", 11)) # to organize into 2 rows
> my_data$pos <- rep(11:1, 2) # specify x coord
>
> p <- ggplot(my_data, aes(x = pos, y = row, size = population)) +
+ geom_point(aes(size = population), shape = 21, fill = "white",
+ show_guide = FALSE) +
+ geom_text(aes(label = city), vjust = 2.7, hjust = 0.95, angle = 40,
+ size = 4) +
+ scale_size_continuous(range = c(3, 12)) +
+ ggtitle("Fig 2.18 Playfair's Population of Cities") +
+ theme_bw() +
+ theme(panel.grid.major = element_blank(),
+ plot.title = element_text(size = rel(1.5), face = "bold", vjust = 1.5),
+ axis.title = element_blank(),
+ axis.text = element_blank(),
+ axis.ticks = element_blank())
Warning message:
`show_guide` has been deprecated. Please use `show.legend` instead.
>
> p
>
> ggsave(paste0(this_base, ".png"),
+ p, width = 7, height = 5)

An Example- Stacked Bar Chart
library(ggplot2)
library(reshape2)
data <- textConnection("Month,Series 1,Series 2,Series 3,Series 4
Jan,7.41,9.38,5.52,6.25
Feb,5.74,8.27,7.29,3.39
Mar,6.52,5.42,7.51,6.20
Apr,2.02,0.70,0.24,1.88
May,7.90,0.35,9.99,6.84
Jun,3.22,8.01,0.91,1.61
Jul,1.43,8.54,8.08,7.62
Aug,9.80,7.79,8.71,8.21
Sep,2.36,8.17,5.70,4.48
Oct,4.39,9.71,7.19,4.96
Nov,3.24,0.26,7.65,1.37
Dec,8.44,7.78,9.44,3.65
")
data <- read.csv(data, h=T)
data$Month <- factor(data$Month, data$Month)
data.lng <- melt(data, id=c("Month"))
p <- ggplot(aes(x=Month, weight=value, fill=variable), data=data.lng)
p + geom_bar() +
coord_flip() +
scale_x_discrete("Legend Title") +
labs(x="X Label", y="Y Label", title="An Example- Stacked Bar Chart prepared by Volkan OBAN")
# full output: http://www.yaksis.com/static/img/03/large/StackedBar.png

line
n <- 60
m <- 50
x <- seq(-4,4, len=m)
# Make up some fake y data
y <- matrix(NA, n, m)
for (i in 1:n) y[i,] <- dnorm(x)*runif(m, 0.5,1)
par(bg="black")
yrange <- range(c(y, y+n/20))
plot(x, x, type="n", axes=FALSE, bg="black", ylim=yrange)
for (i in n:1) {
y1 <- c(y[i,] + i/20, 0, 0)
x1 <- c(x, x[m], x[1])
polygon(x1,y1,col="black")
lines(x, y[i,] + i/20, col="white")
}

ggplot2 example
set.seed(1) # for reproducibility
Day <- c(rep(1:10,each=24))
Hour <- rep(1:24)
data <- data.frame(Day,Hour)
data$Sunlight <- with(data,-10*cos(2*pi*(Hour-1+abs(rnorm(240)))/24))
data$Sunlight[data$Sunlight<0] <- 0
library(ggplot2)
ggplot(data,aes(x=Hour,y=10+24*Day+Hour-1))+
geom_tile(aes(color=Sunlight),size=2)+
scale_color_gradient(low="black",high="yellow")+
ylim(0,250)+ labs(y="",x="")+
coord_polar(theta="x")+
theme(panel.background=element_rect(fill="black"),panel.grid=element_blank(),
axis.text.y=element_blank(), axis.text.x=element_text(color="white"),
axis.ticks.y=element_blank())
library(DiagrammeR)
> library(DiagrammeR)
> library(magrittr)
> graph <-
+ create_graph() %>%
+ set_graph_name("DAG") %>%
+ set_global_graph_attrs("graph", "overlap", "true") %>%
+ set_global_graph_attrs("graph", "fixedsize", "true") %>%
+ set_global_graph_attrs("node", "color", "blue") %>%
+ set_global_graph_attrs("node", "fontname", "Helvetica") %>%
+ add_n_nodes(11) %>%
+ select_nodes_by_id(c(1:4, 8:11)) %>%
+ set_node_attrs_ws("shape", "box") %>%
+ clear_selection %>%
+ select_nodes_by_id(5:7) %>%
+ set_node_attrs_ws("shape", "circle") %>%
+ clear_selection %>%
+ add_edges_w_string(
+ "1->5 2->6 3->9 4->7 5->8 5->10 7->11", "green") %>%
+ add_edges_w_string(
+ "1->8 3->6 3->11 3->7 5->9 6->10", "red") %>%
+ select_edges("rel", "green") %>%
+ set_edge_attrs_ws("color", "green") %>%
+ invert_selection %>%
+ set_edge_attrs_ws("color", "red")
>
> render_graph(graph)
>
ternaryplot
library(vcd)
a<- c (0.1, 0.5, 0.5, 0.6, 0.2, 0, 0, 0.004166667, 0.45)
b<- c (0.75,0.5,0,0.1,0.2,0.951612903,0.918103448,0.7875,0.45)
c<- c (0.15,0,0.5,0.3,0.6,0.048387097,0.081896552,0.208333333,0.1)
d<- c (500,2324.90,2551.44,1244.50, 551.22,-644.20,-377.17,-100, 2493.04)
df<- data.frame (a, b, c)
# First create the limit of the ternary plot:
plot(NA,NA,xlim=c(0,1),ylim=c(0,sqrt(3)/2),asp=1,bty="n",axes=F,xlab="",ylab="")
segments(0,0,0.5,sqrt(3)/2)
segments(0.5,sqrt(3)/2,1,0)
segments(1,0,0,0)
text(0.5,(sqrt(3)/2),"c", pos=3)
text(0,0,"a", pos=1)
text(1,0,"b", pos=1)
# The biggest difficulty in the making of a ternary plot is to transform triangular coordinates into cartesian coordinates, here is a small function to do so:
tern2cart <- function(coord){
coord[1]->x
coord[2]->y
coord[3]->z
x+y+z -> tot
x/tot -> x # First normalize the values of x, y and z
y/tot -> y
z/tot -> z
(2*y + z)/(2*(x+y+z)) -> x1 # Then transform into cartesian coordinates
sqrt(3)*z/(2*(x+y+z)) -> y1
return(c(x1,y1))
}
# Apply this equation to each set of coordinates
t(apply(df,1,tern2cart)) -> tern
# Intrapolate the value to create the contour plot
resolution <- 0.001
require(akima)
interp(tern[,1],tern[,2],z=d, xo=seq(0,1,by=resolution), yo=seq(0,1,by=resolution)) -> tern.grid
# And then plot:
image(tern.grid,breaks=c(-1000,0,500,1000,1500,2000,3000),col=rev(heat.colors(6)),add=T)
contour(tern.grid,levels=c(-1000,0,500,1000,1500,2000,3000),add=T)
points(tern,pch=19)

Sales Funnel visualization with R
library(dplyr)
library(ggplot2)
library(reshape2)
# creating a data samples
# content
df.content <- data.frame(content = c('main', 'ad landing',
'product 1', 'product 2', 'product 3', 'product 4',
'shopping cart',
'thank you page'),
step = c('awareness', 'awareness',
'interest', 'interest', 'interest', 'interest',
'desire',
'action'),
number = c(150000, 80000,
80000, 40000, 35000, 25000,
130000,
120000))
# customers
df.customers <- data.frame(content = c('new', 'engaged', 'loyal'),
step = c('new', 'engaged', 'loyal'),
number = c(25000, 40000, 55000))
# combining two data sets
df.all <- rbind(df.content, df.customers)
# calculating dummies, max and min values of X for plotting
df.all <- df.all %>%
group_by(step) %>%
mutate(totnum = sum(number)) %>%
ungroup() %>%
mutate(dum = (max(totnum) - totnum)/2,
maxx = totnum + dum,
minx = dum)
# data frame for plotting funnel lines
df.lines <- df.all %>%
select(step, maxx, minx) %>%
group_by(step) %>%
unique() %>%
ungroup()
# data frame with dummies
df.dum <- df.all %>%
select(step, dum) %>%
unique() %>%
mutate(content = 'dummy',
number = dum) %>%
select(content, step, number)
# data frame with rates
conv <- df.all$totnum[df.all$step == 'action']
df.rates <- df.all %>%
select(step, totnum) %>%
group_by(step) %>%
unique() %>%
ungroup() %>%
mutate(prevnum = lag(totnum),
rate = ifelse(step == 'new' | step == 'engaged' | step == 'loyal',
round(totnum / conv, 3),
round(totnum / prevnum, 3))) %>%
select(step, rate)
df.rates <- na.omit(df.rates)
# creting final data frame
df.all <- df.all %>%
select(content, step, number)
df.all <- rbind(df.all, df.dum)
df.all <- df.all %>%
group_by(step) %>%
arrange(desc(content)) %>%
ungroup()
# calculating position of labels
df.all <- df.all %>%
group_by(step) %>%
mutate(pos = cumsum(number) - 0.5*number) %>%
ungroup()
# defining order of steps
df.all$step <- factor(df.all$step, levels = c('loyal', 'engaged', 'new', 'action', 'desire', 'interest', 'awareness'))
list <- c(unique(as.character(df.all$content)))
df.all$content <- factor(df.all$content, levels = c('dummy', c(list)))
# creating custom palette with 'white' color for dummies
cols <- c("#ffffff", "#fec44f", "#fc9272", "#a1d99b", "#fee0d2", "#2ca25f",
"#8856a7", "#43a2ca", "#fdbb84", "#e34a33",
"#a6bddb", "#dd1c77", "#ffeda0", "#756bb1")
# plotting chart
ggplot() +
theme_minimal() +
coord_flip() +
scale_fill_manual(values=cols) +
geom_bar(data=df.all, aes(x=step, y=number, fill=content), stat="identity", width=1) +
geom_text(data=df.all[df.all$content!='dummy', ],
aes(x=step, y=pos, label=paste0(content, '-', number/1000, 'K')),
size=4, color='white', fontface="bold") +
geom_ribbon(data=df.lines, aes(x=step, ymax=max(maxx), ymin=maxx, group=1), fill='white') +
geom_line(data=df.lines, aes(x=step, y=maxx, group=1), color='darkred', size=4) +
geom_ribbon(data=df.lines, aes(x=step, ymax=minx, ymin=min(minx), group=1), fill='white') +
geom_line(data=df.lines, aes(x=step, y=minx, group=1), color='darkred', size=4) +
geom_text(data=df.rates, aes(x=step, y=(df.lines$minx[-1]), label=paste0(rate*100, '%')), hjust=1.2,
color='darkblue', fontface="bold") +
theme(legend.position='none', axis.ticks=element_blank(), axis.text.x=element_blank(),
axis.title.x=element_blank())
ref: analyzecore.com/2015/06/23/sales-funnel-visualization-with-r/

Gviz package
install package and reference:
https://bioconductor.org/packages/release/bioc/html/Gviz.html
library(circlize)
> library(circlize)
>
> par(mar = c(1, 1, 1, 1))
> bed1 = generateRandomBed(nr = 100)
> bed1 = bed1[sample(nrow(bed1), 20), ]
> bed2 = generateRandomBed(nr = 100)
> bed2 = bed2[sample(nrow(bed2), 20), ]
> circos.par("track.height" = 0.1, cell.padding = c(0, 0, 0, 0))
> circos.initializeWithIdeogram()
>
> circos.genomicLink(bed1, bed2, col = sample(1:5, 20, replace = TRUE), border = NA)
> circos.clear()
visualize package.
#Evaluates lower tail.
visualize.nbinom(stat = 1, size = 5, prob = 0.5, section = "lower", strict = 0)
#Evaluates bounded region.
visualize.nbinom(stat = c(1,3), size = 10, prob = 0.35, section = "bounded",
strict = c(TRUE, FALSE))
#Evaluates upper tail.
visualize.nbinom(stat = 1, size = 5, prob = 0.5, section = "upper", strict = 1)

visualize package.
#Evaluates lower tail.
visualize.logis(stat = 1, location = 4, scale = 2, section = "lower")
#Evaluates bounded region.
visualize.logis(stat = c(3,5), location = 4, scale = 2, section = "bounded")
#Evaluates upper tail.
visualize.logis(stat = 1, location = 4, scale = 2, section = "upper")

lmtest package
> source("https://www.r-statistics.com/wp-content/uploads/2010/07/coefplot.r.txt")
>
> data("Mroz", package = "car")
> fm <- glm(lfp ~ ., data = Mroz, family = binomial)
> coefplot(fm, parm = -1)

mapmate
library(mapmate)
> library(dplyr)
> library(purrr)
data(annualtemps)
library(RColorBrewer)
pal <- rev(brewer.pal(11, "RdYlBu"))
temps <- mutate(annualtemps, frameID = Year - min(Year) + 1)
frame1 <- filter(temps, frameID == 1) # subset to first frame
save_map(frame1, ortho = FALSE, col = pal, type = "maptiles", save.plot = FALSE,
return.plot = TRUE)
save_map(frame1, col = pal, type = "maptiles", save.plot = FALSE, return.plot = TRUE)

Fractals
library(numDeriv)
library(RColorBrewer)
library(gridExtra)
## Polynom: choose only one or try yourself
f <- function (z) {z^3-1} #Blurry 1
#f <- function (z) {z^4+z-1} #Blurry 2
#f <- function (z) {z^5+z^3+z-1} #Blurry 3
z <- outer(seq(-2, 2, by = 0.01),1i*seq(-2, 2, by = 0.01),'+')
for (k in 1:5) z <- z-f(z)/matrix(grad(f, z), nrow=nrow(z))
## Supressing texts, titles, ticks, background and legend.
opt <- theme(legend.position="none",
panel.background = element_blank(),
axis.ticks=element_blank(),
axis.title=element_blank(),
axis.text =element_blank())
z <- data.frame(expand.grid(x=seq(ncol(z)), y=seq(nrow(z))), z=as.vector(exp(-Mod(f(z)))))
# Create plots. Choose a palette with display.brewer.all()
p1 <- ggplot(z, aes(x=x, y=y, color=z)) + geom_tile() + scale_colour_gradientn(colours=brewer.pal(8, "Paired")) + opt
p2 <- ggplot(z, aes(x=x, y=y, color=z)) + geom_tile() + scale_colour_gradientn(colours=brewer.pal(7, "Paired")) + opt
p3 <- ggplot(z, aes(x=x, y=y, color=z)) + geom_tile() + scale_colour_gradientn(colours=brewer.pal(6, "Paired")) + opt
p4 <- ggplot(z, aes(x=x, y=y, color=z)) + geom_tile() + scale_colour_gradientn(colours=brewer.pal(5, "Paired")) + opt
# Arrange four plots in a 2x2 grid
grid.arrange(p1, p2, p3, p4, ncol=2)

rGreat package
ref:https://github.com/jokergoo/rGREAT

library(gtrellis)
library(gtrellis)
bed = circlize::generateRandomBed()
gtrellis_layout(track_ylim = range(bed[[4]]))
add_track(bed, panel.fun = function(bed) {
x = (bed[[2]] + bed[[3]]) / 2
y = bed[[4]]
grid.points(x, y, pch = 16, size = unit(1, "mm"))
})

yarrr package
code and example:
https://www.r-bloggers.com/the-yarrr-package-0-0-8-is-finally-on-cran/?utm_source=feedburner&utm_medium=email&utm_campaign=Feed%3A+RBloggers+%28R+bloggers%29

yarrr package
library(yarrr)
pirateplot(formula = budget ~ creative.type,
data = subset(movies, budget > 0 &
creative.type %in% c("Multiple Creative Types", "Factual") == FALSE),
point.o = .02,
xlab = "Movie Creative Type",
main = "Movie budgets (in millions) by rating",
gl.col = "gray",
pal = "black")
mtext("-Prepared by Volkan OBAN",
side = 3,
font = 3)
mtext("*Superhero movies tend to have the highest budgets\n...by far!",
side = 1, adj = 1, line = 3,
cex = .8, font = 3)

fmsb package
> maxmin <- data.frame(
+ total=c(5, 1),
+ phys=c(15, 3),
+ psycho=c(3, 0),
+ social=c(5, 1),
+ env=c(5, 1))
> # data for radarchart function version 1 series, minimum value must be omitted from above.
> RNGkind("Mersenne-Twister")
> set.seed(123)
> dat <- data.frame(
+ total=runif(3, 1, 5),
+ phys=rnorm(3, 10, 2),
+ psycho=c(0.5, NA, 3),
+ social=runif(3, 1, 5),
+ env=c(5, 2.5, 4))
> dat <- rbind(maxmin,dat)
> op <- par(mar=c(1, 2, 2, 1),mfrow=c(2, 2))
> radarchart(dat, axistype=1, seg=5, plty=1, vlabels=c("Total\nQOL", "Physical\naspects",
+ "Phychological\naspects", "Social\naspects", "Environmental\naspects"),
+ title="(axis=1, 5 segments, with specified vlabels)", vlcex=0.5)
> radarchart(dat, axistype=2, pcol=topo.colors(3), plty=1, pdensity=c(5, 10, 30),
+ pangle=c(10, 45, 120), pfcol=topo.colors(3),
+ title="(topo.colors, fill, axis=2)")
> radarchart(dat, axistype=3, pty=32, plty=1, axislabcol="grey", na.itp=FALSE,
+ title="(no points, axis=3, na.itp=FALSE)")
> radarchart(dat, axistype=1, plwd=1:5, pcol=1, centerzero=TRUE,
+ seg=4, caxislabels=c("worst", "", "", "", "best"),
+ title="(use lty and lwd but b/w, axis=1,\n centerzero=TRUE, with centerlabels)")
> par(op)
>
fmsb package
>libraryfmsb)
> library(zoo)
>
> dat<-as.data.frame(sunspot.month)
> dat$TS<-seq(as.yearmon("1749-01-01"), as.yearmon("2013-09-01"), by = 1/12)
> colnames(dat)[1] <- "sunspot"
> dat$decade <- floor(as.numeric(format(dat$TS, "%Y"))/10)*10
> dat$century <- floor(as.numeric(format(dat$TS, "%Y"))/100)*100
> dat$month <- format(dat$TS, "%b")
> dat$month <- factor(dat$month, levels = unique(dat$month))
> library(reshape2)
>
> agg <- recast(data = dat,century~month, measure.var = "sunspot", mean)
> MX <- c(NA, rep(max(agg[,-1]), ncol(agg)-1))
> MN <- c(NA, rep(min(agg[,-1]), ncol(agg)-1))
>
> agg <- rbind(MX, MN, agg)
> radarchart(agg[,-1])
circlize package example
library(circlize)
factors = sample(letters[1:6], 100, replace = TRUE)
x = rnorm(100)
y = rnorm(100)
par(mar = c(1, 1, 1, 1))
circos.initialize(factors = factors, x = x)
circos.trackPlotRegion(factors = factors, x = x, y = y, bg.col = "#EEEEEE",
bg.border = NA, track.height = 0.4, panel.fun = function(x, y) {
cell.xlim = get.cell.meta.data("cell.xlim")
cell.ylim = get.cell.meta.data("cell.ylim")
# reference lines
for(xi in seq(cell.xlim[1], cell.xlim[2], length.out = 10)) {
circos.lines(c(xi, xi), cell.ylim, lty = 2, col = "white")
}
for(yi in seq(cell.ylim[1], cell.ylim[2], length.out = 5)) {
circos.lines(cell.xlim, c(yi, yi), lty = 2, col = "white")
}
xlim = get.cell.meta.data("xlim")
ylim = get.cell.meta.data("ylim")
circos.rect(xlim[1], 1, xlim[2], ylim[2], col = "#FF000020", border = NA)
circos.rect(xlim[1], ylim[1], xlim[2], -1, col = "#00FF0020", border = NA)
circos.points(x[y >= 1], y[y >= 1], pch = 16, cex = 0.8, col = "red")
circos.points(x[y <= -1], y[y <= -1], pch = 16, cex = 0.8, col = "green")
circos.points(x[y > -1 & y < 1], y[y > -1 & y < 1], pch = 16, cex = 0.5)
})
circos.clear()
circlize package
library(circlize)
df = read.table(textConnection("
brand_from model_from brand_to model_to
VOLVO s80 BMW 5series
BMW 3series BMW 3series
VOLVO s60 VOLVO s60
VOLVO s60 VOLVO s80
BMW 3series AUDI s4
AUDI a4 BMW 3series
AUDI a5 AUDI a5
"), header = TRUE, stringsAsFactors = FALSE)
brand = c(structure(df$brand_from, names=df$model_from), structure(df$brand_to,names= df$model_to))
brand = brand[!duplicated(names(brand))]
brand = brand[order(brand, names(brand))]
brand_color = structure(2:4, names = unique(brand))
model_color = structure(2:8, names = names(brand))
library(circlize)
gap.degree = do.call("c", lapply(table(brand), function(i) c(rep(2, i-1), 8)))
circos.par(gap.degree = gap.degree)
chordDiagram(df[, c(2, 4)], order = names(brand), grid.col = model_color,
directional = 1, annotationTrack = "grid", preAllocateTracks = list(
list(track.height = 0.02))
)
circos.trackPlotRegion(track.index = 2, panel.fun = function(x, y) {
xlim = get.cell.meta.data("xlim")
ylim = get.cell.meta.data("ylim")
sector.index = get.cell.meta.data("sector.index")
circos.text(mean(xlim), mean(ylim), sector.index, col = "white", cex = 0.6, facing = "inside", niceFacing = TRUE)
}, bg.border = NA)
for(b in unique(brand)) {
model = names(brand[brand == b])
highlight.sector(sector.index = model, track.index = 1, col = brand_color[b],
text = b, text.vjust = -1, niceFacing = TRUE)
}
circos.clear()
circlize package
circlize package in R

pheatmeap
library(pheatmap)
> test = matrix(rnorm(200), 20, 10)
> test[1:10, seq(1, 10, 2)] = test[1:10, seq(1, 10, 2)] + 3
> test[11:20, seq(2, 10, 2)] = test[11:20, seq(2, 10, 2)] + 2
> test[15:20, seq(2, 10, 2)] = test[15:20, seq(2, 10, 2)] + 4
> colnames(test) = paste("Test", 1:10, sep = "")
> rownames(test) = paste("Gene", 1:20, sep = "")
>
> # Generate column annotations
> annotation = data.frame(Var1 = factor(1:10 %% 2 == 0, labels = c("Class1", "Class2")), Var2 = 1:10)
> annotation$Var1 = factor(annotation$Var1, levels = c("Class1", "Class2", "Class3"))
> rownames(annotation) = paste("Test", 1:10, sep = "")
>
> pheatmap(test, annotation = annotation)
> pheatmap(test, annotation = annotation, annotation_legend = FALSE)
> pheatmap(test, annotation = annotation, annotation_legend = FALSE, drop_levels = FALSE)
>
> # Specify colors
> Var1 = c("navy", "darkgreen")
> names(Var1) = c("Class1", "Class2")
> Var2 = c("lightgreen", "navy")
>
> ann_colors = list(Var1 = Var1, Var2 = Var2)
>
> pheatmap(test, annotation = annotation, annotation_colors = ann_colors, main = "Example with all the features")
riverplot package
> edges = data.frame(N1 = paste0(rep(LETTERS[1:4], each = 4), rep(1:5, each = 16)),
+ N2 = paste0(rep(LETTERS[1:4], 4), rep(2:6, each = 16)),
+ Value = runif(80, min = 2, max = 5) * rep(c(1, 0.8, 0.6, 0.4, 0.3), each = 16),
+ stringsAsFactors = F)
>
> edges = edges[sample(c(TRUE, FALSE), nrow(edges), replace = TRUE, prob = c(0.8, 0.2)),]
> nodes = data.frame(ID = unique(c(edges$N1, edges$N2)), stringsAsFactors = FALSE)
> #
> nodes$x = as.integer(substr(nodes$ID, 2, 2))
> nodes$y = as.integer(sapply(substr(nodes$ID, 1, 1), charToRaw)) - 65
> rownames(nodes) = nodes$ID
>library(RColorBrewer)
> palette = paste0(brewer.pal(4, "Set1"), "60")
> styles = lapply(nodes$y, function(n) {
+ list(col = palette[n+1], lty = 0, textcol = "black")
+ })
> names(styles) = nodes$ID
> library(riverplot)
>
> rp <- list(nodes = nodes, edges = edges, styles = styles)
> #
> class(rp) <- c(class(rp), "riverplot")
> plot(rp, plot_area = 0.95, yscale=0.06)
ref:http://www.exegetic.biz/blog/2014/08/plotting-flows-with-riverplot/?utm_source=rss&utm_medium=rss&utm_campaign=plotting-flows-with-riverplot

cloud
cloud(prop.table(Titanic, margin = 1:3),
type = c("p", "h"), strip = strip.custom(strip.names = TRUE),
scales = list(arrows = FALSE, distance = 2), panel.aspect = 0.7,
zlab = "Proportion")[, 1]

fancy 3D histogram
hist3D_fancy<- function(x, y, break.func = c("Sturges", "scott", "FD"), breaks = NULL,
colvar = NULL, col="white", clab=NULL, phi = 5, theta = 25, ...){
# Compute the number of classes for a histogram
break.func <- break.func [1]
if(is.null(breaks)){
x.breaks <- switch(break.func,
Sturges = nclass.Sturges(x),
scott = nclass.scott(x),
FD = nclass.FD(x))
y.breaks <- switch(break.func,
Sturges = nclass.Sturges(y),
scott = nclass.scott(y),
FD = nclass.FD(y))
} else x.breaks <- y.breaks <- breaks
# Cut x and y variables in bins for counting
x.bin <- seq(min(x), max(x), length.out = x.breaks)
y.bin <- seq(min(y), max(y), length.out = y.breaks)
xy <- table(cut(x, x.bin), cut(y, y.bin))
z <- xy
xmid <- 0.5*(x.bin[-1] + x.bin[-length(x.bin)])
ymid <- 0.5*(y.bin[-1] + y.bin[-length(y.bin)])
oldmar <- par("mar")
par (mar = par("mar") + c(0, 0, 0, 2))
hist3D(x = xmid, y = ymid, z = xy, ...,
zlim = c(-max(z)/2, max(z)), zlab = "counts", bty= "g",
phi = phi, theta = theta, shade = 0.2, col = col, border = "black",d = 1, ticktype = "detailed")
scatter3D(x, y,z = rep(-max(z)/2, length.out = length(x)),
colvar = colvar, col = gg.col(100),
add = TRUE, pch = 18, clab = clab,
colkey = list(length = 0.5, width = 0.5,
dist = 0.05, cex.axis = 0.8, cex.clab = 0.8))
par(mar = oldmar)}
hist3D_fancy(quakes$long, quakes$lat, colvar=quakes$depth,breaks =30)
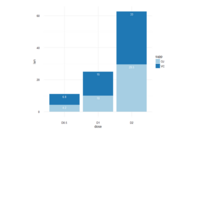
ggplot2 example
df2 <- data.frame(supp=rep(c("VC", "OJ"), each=3),
dose=rep(c("D0.5", "D1", "D2"),2),
len=c(6.8, 15, 33, 4.2, 10, 29.5))
> head(df2)
supp dose len
1 VC D0.5 6.8
2 VC D1 15.0
3 VC D2 33.0
4 OJ D0.5 4.2
5 OJ D1 10.0
6 OJ D2 29.5
> library(plyr)
>
> df_sorted <- arrange(df2, dose, supp)
> head(df_sorted)
> df_cumsum <- ddply(df_sorted, "dose",
transform, label_ypos=cumsum(len))
> ggplot(data=df_cumsum, aes(x=dose, y=len, fill=supp)) +
geom_bar(stat="identity")+
geom_text(aes(y=label_ypos, label=len), vjust=1.6,
color="white", size=3.5)+
scale_fill_brewer(palette="Paired")+
theme_minimal()

ggplot2
> df <- data.frame(supp=rep(c("VC", "OJ"), each=3),
dose=rep(c("D0.5", "D1", "D2"),2),
len=c(6.8, 15, 33, 4.2, 10, 29.5))
> ggplot(data=df, aes(x=dose, y=len, fill=supp)) +
geom_bar(stat="identity", position=position_dodge())+
geom_text(aes(label=len), vjust=1.6, color="white",
position = position_dodge(0.9), size=3.5)+
scale_fill_brewer(palette="Paired")+
theme_minimal()

library(rLiDAR)
> library(rLiDAR)
> data(chm)

treemapify
>library(treemapify)
> library(ggplot2)
> country <- c("Ireland","England","France","Germany","USA","Spain")
> job <- c("IT","SOCIAL","Project Manager","Director","Vice-President")
>
> mydf <- data.frame(countries = sample(country,100,replace = TRUE),
+ career = sample(job,100,replace=TRUE),
+ participent = sample(1:100, replace = TRUE)
+ )
>
> # Set Up the coords
> treemap_coords <- treemapify(mydf,
+ area="participent",
+ fill="countries",
+ label="career",
+ group="countries")
>
> # Plot the results using the Green Pallete
> ggplotify(treemap_coords,
+ group.label.size.factor = 2,
+ group.label.colour = "white",
+ label.colour = "black",
+ label.size.factor = 1) +
+ labs(title="Work Breakdown") +
+ scale_colour_brewer(palette = "Greens")

treemap
treemap(business,
index=c("NACE1", "NACE2", "NACE3"),
vSize="turnover",
type="index")

treemap
library(treemap)
> data(GNI2014)
> treemap(GNI2014,
index=c("continent", "iso3"),
vSize="population",
vColor="GNI", type="value"

dggridR: Discrete Global Grids for R
countries <- map_data("world")
#Plot everything on a flat map
p<- ggplot() +
geom_polygon(data=countries, aes(x=long, y=lat, group=group), fill=NA, color="black") +
geom_polygon(data=grid, aes(x=long, y=lat, group=group), fill="green", alpha=0.4) +
geom_path (data=grid, aes(x=long, y=lat, group=group), alpha=0.4, color="white")
p

dggridR: Discrete Global Grids for R
library(dggridR)
library(dplyr)
data(dgquakes)
p<- ggplot() +
geom_polygon(data=countries, aes(x=long, y=lat, group=group), fill=NA, color="black") +
geom_polygon(data=grid, aes(x=long, y=lat, group=group, fill=count), alpha=0.4) +
geom_path (data=grid, aes(x=long, y=lat, group=group), alpha=0.4, color="white") +
scale_fill_gradient(low="blue", high="red")
p+coord_map("ortho", orientation = c(-38.49831, -179.9223, 0))+
xlab('')+ylab('')+
theme(axis.ticks.x=element_blank())+
theme(axis.ticks.y=element_blank())+
theme(axis.text.x=element_blank())+
theme(axis.text.y=element_blank())+
ggtitle('Your data could look like this')

library(dggridR)
library(dggridR)
d> library(dplyr)
d> dggs <- dgconstruct(spacing=1000, metric=FALSE, resround='down')
d> data(dgquakes)
d> dgquakes$cell <- dgtransform(dggs,dgquakes$lat,dgquakes$lon)
d> quakecounts <- dgquakes %>% group_by(cell) %>% summarise(count=n())
grid <- dgcellstogrid(dggs,quakecounts$cell,frame=TRUE,wrapcells=TRUE)
grid <- merge(grid,quakecounts,by.x="Name",by.y="cell")
d> grid$count <- log(grid$count)
d> cutoff <- quantile(grid$count,0.9)
d> grid <- grid %>% mutate(count=ifelse(count>cutoff,cutoff,count))
d>
d> #Get polygons for each country of the world
d> countries <- map_data("world")
d> p<- ggplot() +
+ geom_polygon(data=countries, aes(x=long, y=lat, group=group), fill=NA, color="black") +
+ geom_polygon(data=grid, aes(x=long, y=lat, group=group, fill=count), alpha=0.4) +
+ geom_path (data=grid, aes(x=long, y=lat, group=group), alpha=0.4, color="white") +
+ scale_fill_gradient(low="blue", high="red")
d> p
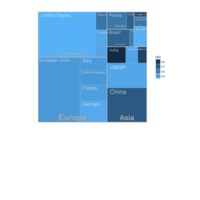
library(treemapify)
>library(treemapify)
> data(G20)
> treeMapCoordinates <- treemapify(G20,
+ area = "Nom.GDP.mil.USD",
+ fill = "HDI",
+ label = "Country",
+ group = "Region")
> treeMapPlot <- ggplotify(treeMapCoordinates)
> print(treeMapPlot)

library(vcd)
> library(vcd)
> mosaic(HairEyeColor, shade=TRUE, legend=TRUE)

wordcloud
> library(wordcloud)
> library(tm)
wordcloud(c(" TURKEY","OBAN","DATA SCIENCE","ANALYTICS","MATHEMATICS","Machine Learnings","Istanbul","Researcher","Philosophy","Mathematician","VOLKAN","Data"),freq = c(25,22,10,24,30,25,7,9,12,18,62,14),min.freq = 0,col="purple")
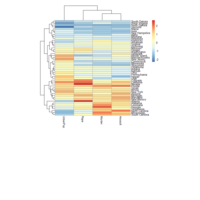
pheatmap
> library(pheatmap)
> data=as.matrix(scale(USArrests))
> clst=hclust(dist(data))
> pheatmap(data)
lattice
library(lattice)
> data("quakes")
> quakes$Magnitude<-equal.count(quakes$mag,4)
> cloud(depth~lat*long | Magnitude, data=quakes, zlim=rev(range(quakes$depth)),screen=list(z=105,x=-70),panel.aspect =0.75,lab="Longitude",ylab = "Latitude",zlab="Depth" )

lattice package
> library(lattice)
> parallel(~mtcars[c(1,3,4,5,6,7)] | factor(cyl),mtcars,groups = carb, layout=c(3,1),auto.key = list(space="top",columns=3))

library(RColorBrewer)
> library(RColorBrewer)
> par(mar = c(0, 4, 0, 0))
> display.brewer.all()
plotly 2D contour plot
library(plotly)
x <- rnorm(200)
y <- rnorm(200)
s <- subplot(
plot_ly(x = x, type = "histogram"),
plotly_empty(),
plot_ly(x = x, y = y, type = "histogram2dcontour"),
plot_ly(y = y, type = "histogram"),
nrows = 2, heights = c(0.2, 0.8), widths = c(0.8, 0.2), margin = 0,
shareX = TRUE, shareY = TRUE, titleX = FALSE, titleY = FALSE
)
layout(s, showlegend = FALSE)
Contour Plots with R
res = contourLines(volcano)
contour(volcano, col = "darkgreen", lwd = 2)
library("igraph")
> d <- read.csv("sociogram-employees-un.csv", header=FALSE)
> g <- graph.adjacency(as.matrix(d), mode="directed")
> V(g)$name <- LETTERS[1:NCOL(d)]
> V(g)$color <- "yellow"
> V(g)$shape <- "sphere"
> E(g)$color <- "gray"
> E(g)$arrow.size <- 0.2
> plot(g)
> diameter.nodes <- get.diameter(g)
> diameter.nodes
+ 4/25 vertices, named:
[1] S B A T
> ## + 4/25 vertices, named:
> ## [1] S B A T
> V(g)$size <- 20
> V(g)[diameter.nodes]$color <- "red"
> V(g)[diameter.nodes]$size <- V(g)[diameter.nodes]$size+10
> E(g)$width <- 1
> E(g, path=diameter.nodes)$color <- "red"
> E(g, path=diameter.nodes)$width <- 2
> plot.igraph(g, layout=layout.fruchterman.reingold)

misc3d
> drawScene(surfaceTriangles(seq(-1,1,len=30), seq(-1,1,len=30),function(x, y) (x^2 + y^2), color2 = "white"))
> drawScene.rgl(surfaceTriangles(seq(-1,1,len=30), seq(-1,1,len=30), function(x, y) (x^2 + y^2), color2 = "white"))
xkcd package in R
library(xkcd)
datascaled <- data.frame(x=c(-3,3),y=c(-30,30))
p <- ggplot(data=datascaled, aes(x=x,y=y)) + geom_point()
xrange <- range(datascaled$x)
yrange <- range(datascaled$y)
ratioxy <- diff(xrange) / diff(yrange)
mapping <- aes(x=x,
y=y,
scale=scale,
ratioxy=ratioxy,
angleofspine = angleofspine,
anglerighthumerus = anglerighthumerus,
anglelefthumerus = anglelefthumerus,
anglerightradius = anglerightradius,
angleleftradius = angleleftradius,
anglerightleg = anglerightleg,
angleleftleg = angleleftleg,
angleofneck = angleofneck,
color = color )
dataman <- data.frame( x= c(-1,0,1), y=c(-10,0,10),
scale = c(10,7,5),
ratioxy = ratioxy,
angleofspine = seq(- pi / 2, -pi/2 + pi/8, l=3) ,
anglerighthumerus = -pi/6,
anglelefthumerus = pi + pi/6,
anglerightradius = 0,
angleleftradius = runif(3,- pi/4, pi/4),
angleleftleg = 3*pi/2 + pi / 12 ,
anglerightleg = 3*pi/2 - pi / 12,
angleofneck = runif(3, min = 3 * pi / 2 - pi/10 , max = 3 * pi / 2 + pi/10),
color=c("A","B","C"))
p + xkcdman(mapping,dataman)

library(d3heatmap)
library(d3heatmap)
d3heatmap(mtcars, scale="column", colors="Blues")

tableplot
tableplot(diamonds, select = 1:7, fontsize = 14, legend.lines = 8, title = "Shine on you crazy Diamond", fontsize.title = 18)

tableplot
tableplot(diamonds, pals = list(cut="Set1(6)", color="Set5", clarity=rainbow(8)))
diamonds$carat_class <- num2fac(diamonds$carat, n=20)
diamonds$price_class <- num2fac(diamonds$price, n=100)
tableplot(diamonds, select=c(carat, price, carat_class, price_class))

coplot
#I'll use the example in the R cookbook
data(Cars93, package="MASS")
coplot(Horsepower ~ MPG.city | Origin, data=Cars93)

tableplot
library(ggplot2)
data(diamonds)
#run ?diamonds for more information on the dataset
tableplot(diamonds)
#sort by depth
tableplot(diamonds, sortCol=depth)

tableplot package
library(tabplot)
#how the iris dataset looks
row.sample <- function(dta, rep) {
dta[sample(1:nrow(dta), rep, replace=FALSE), ]
}
head(row.sample(iris))
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
18 5.1 3.5 1.4 0.3 setosa
71 5.9 3.2 4.8 1.8 versicolor
83 5.8 2.7 3.9 1.2 versicolor
133 6.4 2.8 5.6 2.2 virginica
21 5.4 3.4 1.7 0.2 setosa
144 6.8 3.2 5.9 2.3 virginica
tableplot(iris, sortCol="Species")

data visulazition in R
require(ggplot2)
data(diamonds)
## add some NA's
is.na(diamonds$price) <- diamonds$cut == "Ideal"
is.na(diamonds$cut) <- (runif(nrow(diamonds)) > 0.8)

library(caret)
3
4
5
6
7
8
9
# load the library
library(caret)
# load the data
data(iris)
# density plots for each attribute by class value
x <- iris[,1:4]
y <- iris[,5]
scales <- list(x=list(relation="free"), y=list(relation="free"))
featurePlot(x=x, y=y, plot="density", scales=scales)

library(mlbench)
3
4
5
6
7
8
9
10
11
# load the library
library(mlbench)
# load the dataset
data(BreastCancer)
# create a bar plot of each categorical attribute
par(mfrow=c(2,4))
for(i in 2:9) {
counts <- table(BreastCancer[,i])
name <- names(BreastCancer)[i]
barplot(counts, main=name)
}
timevis and shiny
library(timevis)
timevis()
timevis(
data.frame(id = 1:2,
content = c("one", "two"),
start = c("2016-01-10", "2016-01-12"))
)
#----------------------- Hide the zoom buttons, allow items to be editable -----------------
timevis(
data.frame(id = 1:2,
content = c("one", "two"),
start = c("2016-01-10", "2016-01-12")),
showZoom = FALSE,
options = list(editable = TRUE, height = "200px")
)
#----------------------- You can use %>% pipes to create timevis pipelines -----------------
timevis() %>%
setItems(data.frame(
id = 1:2,
content = c("one", "two"),
start = c("2016-01-10", "2016-01-12")
)) %>%
setOptions(list(editable = TRUE)) %>%
addItem(list(id = 3, content = "three", start = "2016-01-11")) %>%
setSelection("3") %>%
fitWindow(list(animation = FALSE))
#------- Items can be a single point or a range, and can contain HTML -------
timevis(
data.frame(id = 1:2,
content = c("one", "two<br><h3>HTML is supported</h3>"),
start = c("2016-01-10", "2016-01-18"),
end = c("2016-01-14", NA),
style = c(NA, "color: red;")
)
)
#----------------------- Alternative look for each item -----------------
timevis(
data.frame(id = 1:2,
content = c("one", "two"),
start = c("2016-01-10", "2016-01-14"),
end = c(NA, "2016-01-18"),
type = c("point", "background"))
)
#----------------------- Using a function in the configuration options -----------------
timevis(
data.frame(id = 1,
content = "double click anywhere<br>in the timeline<br>to add an item",
start = "2016-01-01"),
options = list(
editable = TRUE,
onAdd = htmlwidgets::JS('function(item, callback) {
item.content = "Hello!<br/>" + item.content;
callback(item);
}')
)
)
#----------------------- Using groups -----------------
timevis(data = data.frame(
start = c(Sys.Date(), Sys.Date(), Sys.Date() + 1, Sys.Date() + 2),
content = c("one", "two", "three", "four"),
group = c(1, 2, 1, 2)),
groups = data.frame(id = 1:2, content = c("G1", "G2"))
)
#----------------------- Getting data out of the timeline into Shiny -----------------
if (interactive()) {
library(shiny)
data <- data.frame(
id = 1:3,
start = c("2015-04-04", "2015-04-05 11:00:00", "2015-04-06 15:00:00"),
end = c("2015-04-08", NA, NA),
content = c("<h2>Vacation!!!</h2>", "Acupuncture", "Massage"),
style = c("color: red;", NA, NA)
)
ui <- fluidPage(
timevisOutput("appts"),
div("Selected items:", textOutput("selected", inline = TRUE)),
div("Visible window:", textOutput("window", inline = TRUE)),
tableOutput("table")
)
server <- function(input, output) {
output$appts <- renderTimevis(
timevis(
data,
options = list(editable = TRUE, multiselect = TRUE, align = "center")
)
)
output$selected <- renderText(
paste(input$appts_selected, collapse = " ")
)
output$window <- renderText(
paste(input$appts_window[1], "to", input$appts_window[2])
)
output$table <- renderTable(
input$appts_data
)
}
shinyApp(ui, server)
}

library("wordcloud")
Cahit Sıtkı Tarancı-Otuz beş Yaş Şiiri

wordcloud
> res<-rquery.wordcloud(filePath, type ="file",colorPalette = "red", lang = "english")

wordcloud
res<-rquery.wordcloud(filePath, type ="file",colorPalette = "blue", lang = "english")

wordcloud
library(wordcloud)
library("tm")
library("SnowballC")
library("wordcloud")
library("RColorBrewer")
filePath <- "C:/Users/lenovo/Documents/text/corpus.txt"
res<-rquery.wordcloud(filePath, type ="file",colorPalette = "black", lang = "english")
qcc package;library(qcc)
> library(qcc)
> x <- rep(10, 100) + rnorm(100)
> new.x <- rep(11, 15) + rnorm(15)
> qcc(x, newdata=new.x, type="xbar.one")

> library(ggplot2)
> library(ggplot2)
> data("iris")
>
> boxplot(Sepal.Width ~ Species, data=iris, ylim=c(2, 4.5), xaxt='n', yaxt='n',notch=TRUE, boxwex=0.5, boxcol="blue", medcol="red", medlwd=1,outcol="red", outpch=3, outcex=0.8)
> boxplot(Sepal.Width ~ Species, data=iris, ylim=c(2, 4.5), xaxt='n', yaxt='n',notch=TRUE, boxwex=0.5, boxcol="blue", medcol="red", medlwd=1,outcol="red", outpch=3, outcex=0.8)
> mtext("Comparison of three species in the Fisher iris data", 3, cex=0.9)
> mtext("Sepal width in mm", 2, cex=0.9, line=2)
> boxplot(Sepal.Width ~ Species, data=iris, ylim=c(2, 4.5), xaxt='n', yaxt='n',notch=TRUE, boxwex=0.5, boxcol="blue", medcol="red", medlwd=1, outcol="red", outpch=3, outcex=0.8)
> mtext("Comparison of three species in the Fisher iris data", 3, cex=0.9)
> mtext("Sepal width in mm", 2, cex=0.9, line=2)
>
> lab <- format(as.character(pretty(c(2,4.5))), drop0trailing=TRUE, justify="right")
> axis(2, tck=0.02, at=pretty(c(2,4.5)), labels=lab, las=1, hadj=0.3)
> axis(4, tck=0.02, labels=FALSE)
> axis(1, at=1:3, labels=unique(iris$Species), tck=0, padj=-1)
library(geomorph)
> data(scallops)
> Y.gpa<-gpagen(A=scallops$coorddata, curves=scallops$curvslide,
+ surfaces=scallops$surfslide)
|=======================================================| 100%
> ref<-mshape(Y.gpa$coords)
> plotRefToTarget(ref,Y.gpa$coords[,,1],method="TPS", mag=3)> data(scallops)
> Y.gpa<-gpagen(A=scallops$coorddata, curves=scallops$curvslide,surfaces=scallops$surfslide)
> ref<-mshape(Y.gpa$coords)
> plotRefToTarget(ref,Y.gpa$coords[,,1],method="TPS", mag=3)
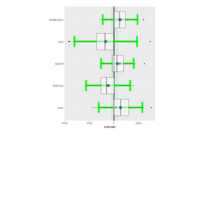
library(PSAboot)
> data(lalonde, package = "MatchIt")
> table(lalonde$treat)
> lalonde.formu <- treat ~ age + I(age^2) + educ + I(educ^2) + black + hispan + married + nodegree +
+ re74 + I(re74^2) + re75 + I(re75^2) + re74 + re75
> boot.lalonde <- PSAboot(Tr = lalonde$treat, Y = lalonde$re78, X = lalonde, formu = lalonde.formu,
+ M = 100, seed = 2112)
boxplot(boot.lalonde)

library(PSAboot)
data(lalonde, package = "MatchIt")
> lalonde.formu <- treat ~ age + I(age^2) + educ + I(educ^2) + black + hispan + married + nodegree +
+ re74 + I(re74^2) + re75 + I(re75^2) + re74 + re75
> boot.lalonde <- PSAboot(Tr = lalonde$treat, Y = lalonde$re78, X = lalonde, formu = lalonde.formu,
+ M = 100, seed = 2112)
plot(boot.lalonde)
plotly-subplot
> library(plotly)
> p <- subplot(
+ plot_ly(economics, x = date, y = uempmed),
+ plot_ly(economics, x = date, y = unemploy),
+ margin = 0.05,
+ nrows=2
+ ) %>% layout(showlegend = FALSE)
> p
plotly-subplot
> library(plotly)
> p <- subplot(
+ plot_ly(economics, x = date, y = uempmed),
+ plot_ly(economics, x = date, y = unemploy),
+ margin = 0.05
+ ) %>% layout(showlegend = FALSE)
> p

library(ggthemes)
>library(ggthemes)
> ggplot(nmmaps, aes(date, temp, color=factor(season)))+
+ geom_point()+ggtitle("This plot looks a lot different from the default")+
+ theme_economist()+scale_colour_economist()
ggtree
>library("ggtree")
>data(chiroptera)
> groupInfo <- split(chiroptera$tip.label, gsub("_\\w+", "", chiroptera$tip.label))
> chiroptera <- groupOTU(chiroptera, groupInfo)
> ggtree(chiroptera, aes(color=group), layout='circular') + geom_tiplab(size=1, aes(angle=angle))
>
ggtree
library("ggtree")
nwk <- system.file("extdata", "sample.nwk", package="ggtree")
tree <- read.tree(nwk)
ggplot(tree, aes(x, y)) + geom_tree() + theme_tree()

ggtree
ggtree
https://bioconductor.org/packages/release/bioc/html/ggtree.html
library(ggplot2)
library(ape)
library(ggtree)
file <- system.file("extdata/BEAST", "beast_mcc.tree", package="ggtree")
beast <- read.beast(file)
ggtree(beast, ndigits=2, branch.length = 'none') + geom_text(aes(x=branch, label=length_0.95_HPD), vjust=-.5, color='firebrick')

Waffleplot
tiles <- c(One=80, Two=30, Three=20, Four=10)
Waffleplot(tiles, rows=8)
Senate <- c(`Male (44%)`=44, `Female (56%)`=56)
Waffleplot(Senate, rows=10, size=0.5, colors=c("#af9139", "#544616"))
Sciencepro package
library(sciencepro)
parts <- c(80, 30, 20, 10)
> w1 <- Waffleplot(parts, rows=8)
> w2 <- Waffleplot(parts, rows=8)
> w3 <- Waffleplot(parts, rows=8)
> chart <- Forge(w1, w2, w3)
> print(chart)

ggalt
> world <- map_data("world")
>
> world <- world[world$region != "Antarctica",]
>
> gg <- ggplot()
> gg <- gg + geom_map(data=world, map=world,
+ aes(x=long, y=lat, map_id=region))
> gg <- gg + coord_proj("+proj=wintri")
> gg
ggalt-geom_dumbbell
> library(tidyr)
> library(scales)
> library(ggplot2)
> library(ggalt) # devtools::install_github("hrbrmstr/ggalt")
>
> health <- read.csv("https://gist.githubusercontent.com/hrbrmstr/0d206070cea01bcb0118/raw/0ea32190a8b2f54b5a9770cb6582007132571c98/zhealth.csv", stringsAsFactors=FALSE,
+ header=FALSE, col.names=c("pct", "area_id"))
>
> areas <- read.csv("https://gist.githubusercontent.com/hrbrmstr/0d206070cea01bcb0118/raw/0ea32190a8b2f54b5a9770cb6582007132571c98/zarea_trans.csv", stringsAsFactors=FALSE, header=TRUE)
>
> health %>%
+ mutate(area_id=trunc(area_id)) %>%
+ arrange(area_id, pct) %>%
+ mutate(year=rep(c("2014", "2013"), 26),
+ pct=pct/100) %>%
+ left_join(areas, "area_id") %>%
+ mutate(area_name=factor(area_name, levels=unique(area_name))) -> health
>
> setNames(bind_cols(filter(health, year==2014), filter(health, year==2013))[,c(4,1,5)],
+ c("area_name", "pct_2014", "pct_2013")) -> health
>
>
> gg <- ggplot(health, aes(x=pct_2013, xend=pct_2014, y=area_name, group=area_name))
> gg <- gg + geom_dumbbell(color="#a3c4dc", size=0.75, point.colour.l="#0e668b")
> gg <- gg + scale_x_continuous(label=percent)
> gg <- gg + labs(x=NULL, y=NULL)
> gg <- gg + theme_bw()
> gg <- gg + theme(plot.background=element_rect(fill="#f7f7f7"))
> gg <- gg + theme(panel.background=element_rect(fill="#f7f7f7"))
> gg <- gg + theme(panel.grid.minor=element_blank())
> gg <- gg + theme(panel.grid.major.y=element_blank())
> gg <- gg + theme(panel.grid.major.x=element_line())
> gg <- gg + theme(axis.ticks=element_blank())
> gg <- gg + theme(legend.position="top")
> gg <- gg + theme(panel.border=element_blank())
> gg
Lollipop charts-geom_lollipop() by the Chartettes
df <- read.csv(text="category,pct
Other,0.09
South Asian/South Asian Americans,0.12
Interngenerational/Generational,0.21
S Asian/Asian Americans,0.25
Muslim Observance,0.29
Africa/Pan Africa/African Americans,0.34
Gender Equity,0.34
Disability Advocacy,0.49
European/European Americans,0.52
Veteran,0.54
Pacific Islander/Pacific Islander Americans,0.59
Non-Traditional Students,0.61
Religious Equity,0.64
Caribbean/Caribbean Americans,0.67
Latino/Latina,0.69
Middle Eastern Heritages and Traditions,0.73
Trans-racial Adoptee/Parent,0.76
LBGTQ/Ally,0.79
Mixed Race,0.80
Jewish Heritage/Observance,0.85
International Students,0.87", stringsAsFactors=FALSE, sep=",", header=TRUE)
library(ggplot2)
library(ggalt)
library(scales)
gg <- ggplot(df, aes(y=reorder(category, pct), x=pct))
gg <- gg + geom_lollipop(point.colour="steelblue", point.size=3, horizontal=TRUE)
gg <- gg + scale_x_continuous(expand=c(0,0), labels=percent,
breaks=seq(0, 1, by=0.2), limits=c(0, 1))
gg <- gg + labs(x=NULL, y=NULL,
title="SUNY Cortland Multicultural Alumni survey results",
subtitle="Ranked by race, ethnicity, home land and orientation\namong the top areas of concern",
caption="Data from http://stephanieevergreen.com/lollipop/")
gg <- gg + theme_minimal(base_family="Arial Narrow")
gg <- gg + theme(panel.grid.major.y=element_blank())
gg <- gg + theme(panel.grid.minor=element_blank())
gg <- gg + theme(axis.line.y=element_line(color="#2b2b2b", size=0.15))
gg <- gg + theme(axis.text.y=element_text(margin=margin(r=-5, l=0)))
gg <- gg + theme(plot.margin=unit(rep(30, 4), "pt"))
gg <- gg + theme(plot.title=element_text(face="bold"))
gg <- gg + theme(plot.subtitle=element_text(margin=margin(b=10)))
gg <- gg + theme(plot.caption=element_text(size=8, margin=margin(t=10)))
gg

ggalt
m + stat_bkde2d(bandwidth=c(0.5, 4), aes(fill = ..level..), geom = "polygon")
Alternate 2D density plots-library(ggplot2) library(gridExtra) library(ggalt)
m <- ggplot(faithful, aes(x = eruptions, y = waiting)) +
+ geom_point() +
+ xlim(0.5, 6) +
+ ylim(40, 110)
There were 12 warnings (use warnings() to see them)
>
> m + geom_bkde2d(bandwidth=c(0.5, 4))
library(maps)
map('world',proj='orth',orient=c(41,-74,0))

ggrepel
p <- p + theme_tufte() + theme(
axis.title.y = element_text(vjust=1, angle=0, hjust=1),
legend.position='none')
p

ggrepel
> p <- p + scale_colour_manual(values = hiva.contrast) +
+ scale_fill_manual(values = hiva.contrast)
>
> # add titles and annotation for the median lines
> p <- p + labs(
+ title='Labor in the transportation sector is on average some of the most\n strained work: high intensity, low autonomy (Eurofound, EWCS 2010)',
+ x='Work intensity', y='Job\nautonomy')
>
> p <- p + annotate('text',
+ label='EU median',
+ x=34.5,y=43, color=hiva.grijs)
>
> p <- p + annotate('text',
+ label='EU median',
+ x=26,y=59, color=hiva.grijs)
>
> p

ggrepel
> library(ggplot2)
> library(ggrepel)
> library(ggthemes)
> strain.sector <- read.csv2('https://git.io/v2lGL')
> dim(strain.sector)
[1] 32 3
> strain.sector$highlight <- ifelse(
+ strain.sector$sector_label == 'Transport and storage',
+ TRUE, FALSE)
> hiva.oranje.donker <- '#F67504'
> hiva.oranje.licht <- "#fc9e49" # officieel
> hiva.grijs <- 'grey60'
> hiva.groen.licht <- "#bad80a"
> hiva.groen.donker <- "#8EA608"
> hiva.contrast <- c(hiva.grijs, hiva.oranje.donker)
> p <- ggplot(strain.sector, aes(
+ x=work_intensity,
+ y=job_autonomy,
+ group=highlight,
+ color=highlight))
> p <- p + geom_hline(yintercept=60, colour=hiva.grijs) +
+ geom_vline(xintercept=37, colour=hiva.grijs)
>
> # Add points
> p <- p + geom_point(size = 5, color='grey80')
> p
> set.seed(42) # set a reed to get the same label-placement
> p <- p + geom_label_repel(
+ aes(
+ fill = highlight,
+ label = sector_label),
+ fontface = 'bold', color = 'white',
+ size = 2,
+ box.padding = unit(0.25, "lines"),
+ point.padding = unit(0.5, "lines")
+ )
> p

ggrepel
library(ggrepel)
set.seed(42)
> ggplot(mtcars) +
+ geom_point(aes(wt, mpg), color = 'grey', size = 4, shape = 15) +
+ geom_text_repel(
+ aes(
+ wt, mpg,
+ color = factor(cyl),
+ label = rownames(mtcars)
+ ),
+ size = 5,
+ fontface = 'bold',
+ box.padding = unit(0.5, 'lines'),
+ point.padding = unit(1.6, 'lines'),
+ segment.color = '#555555',
+ segment.size = 0.5,
+ arrow = arrow(length = unit(0.01, 'npc')),
+ force = 1,
+ max.iter = 2e3,
+ nudge_x = ifelse(mtcars$cyl == 6, 1, 0),
+ nudge_y = ifelse(mtcars$cyl == 6, 8, 0)
+ ) +
+ scale_color_discrete(name = 'cyl') +
+ theme_classic(base_size = 16)
>

library(ggrepel)
> library(ggplot2)
> library(ggrepel)
> ggplot(mtcars, aes(wt, mpg)) +
+ geom_point(color = 'red') +
+ geom_text_repel(aes(label = rownames(mtcars))) +
+ theme_classic(base_size = 16)
ggiraph
g <- ggplot(mpg, aes( x = displ, y = cty, color = drv) ) + theme_minimal()
my_gg <- g + geom_point_interactive(aes(tooltip = model), size = 2)
ggiraph(code = print(my_gg), width = .7)

geomnet
library(ggplot2)
data(blood)
p <- ggplot(data = blood$edges, aes(from_id = from, to_id = to))
p + geom_net(vertices=blood$vertices, aes(colour=..type..)) + theme_net()
bloodnet <- merge(blood$edges, blood$vertices, by.x="from", by.y="label", all=TRUE)
p <- ggplot(data = bloodnet, aes(from_id = from, to_id = to))
p + geom_net()
p + geom_net(aes(colour=rho)) + theme_net()
p + geom_net(aes(colour=rho), label=TRUE, vjust = -0.5)
p + geom_net(aes(colour=rho), label=TRUE, vjust=-0.5, labelcolour="black",
directed=TRUE, curvature=0.2) + theme_net()
p + geom_net(colour = "orange", layout = 'circle', size = 6)
p + geom_net(colour = "orange", layout = 'circle', size = 6, linewidth=.75)
p + geom_net(colour = "orange", layout = 'circle', size = 0, linewidth=.75,
directed = TRUE)
p + geom_net(aes(size=Predominance, colour=rho, shape=rho, linetype=group_to),
linewidth=0.75, label =TRUE, labelcolour="black") +
facet_wrap(~Ethnicity) +
scale_colour_brewer(palette="Set2")
gg <- ggplot(data = blood$edges, aes(from_id = from, to_id = to)) +
geom_net(colour = "darkred", layout = "circle", label = TRUE, size = 15,
directed = TRUE, vjust = 0.5, labelcolour = "grey80",
arrowsize = 1.5, linewidth = 0.5, arrowgap = 0.05,
selfies = TRUE, ecolour = "grey40") +
theme_net()
gg
dframe <- ggplot_build(gg)$data[[1]] # contains calculated node and edge values
10 geom_net
#Madmen Relationships
data(madmen)
MMnet <- merge(madmen$edges, madmen$vertices, by.x="Name1", by.y="label", all=TRUE)
p <- ggplot(data = MMnet, aes(from_id = Name1, to_id = Name2))
p + geom_net(label=TRUE)
p + geom_net(aes(colour=Gender), size=6, linewidth=1, label=TRUE, fontsize=3, labelcolour="black")
p + geom_net(aes(colour=Gender), size=6, linewidth=1, label=TRUE, labelcolour="black") +
scale_colour_manual(values=c("#FF69B4", "#0099ff")) + xlim(c(-.05,1.05))
p + geom_net(aes(colour=Gender), size=6, linewidth=1, directed=TRUE, label=TRUE,
arrowgap=0.01, labelcolour="black") +
scale_colour_manual(values=c("#FF69B4", "#0099ff")) + xlim(c(-.05,1.05))
p <- ggplot(data = MMnet, aes(from_id = Name1, to_id = Name2))
# alternative labelling: specify label variable.
p + geom_net(aes(colour=Gender, label=Gender), size=6, linewidth=1, fontsize=3,
labelcolour="black")
## visualizing ggplot2 theme elements
data(theme_elements)
TEnet <- merge(theme_elements$edges, theme_elements$vertices, by.x="parent",
by.y="name", all=TRUE)
ggplot(data = TEnet, aes(from_id = parent, to_id = child)) +
geom_net(label=TRUE, vjust=-0.5)
## emails example from VastChallenge 2014
# care has to be taken to make sure that for each panel all nodes are included with
# the necessary information.
# Otherwise line segments show on the plot without nodes.
data(email)
employee <- data.frame(expand.grid(
label=unique(email$nodes$label), day=unique(email$edges$day)))
employee <- merge(employee, email$nodes, by="label")
emailnet <- merge(subset(email$edges, nrecipients < 54), employee,
by.x=c("From", "day"), by.y=c("label", "day"), all=TRUE)
#no facets
ggplot(data = emailnet, aes(from_id = From, to_id = to)) +
geom_net(aes(colour= CurrentEmploymentType), linewidth=0.5) +
scale_colour_brewer(palette="Set2")
#facet by day
ggplot(data = emailnet, aes(from_id = From, to_id = to)) +
geom_net(aes(colour= CurrentEmploymentType), linewidth=0.5, fiteach=TRUE) +
scale_colour_brewer(palette="Set2") +
facet_wrap(~day, nrow=2) + theme(legend.position="bottom")
ggplot(data = emailnet, aes(from_id = From, to_id = to)) +
geom_net(aes(colour= CitizenshipCountry), linewidth=0.5, fiteach=TRUE) +
scale_colour_brewer(palette="Set2") +
facet_wrap(~day, nrow=2) + theme(legend.position="bottom")
ggplot(data = emailnet, aes(from_id = From, to_id = to)) +
geom_net(aes(colour= CurrentEmploymentType), linewidth=0.5, fiteach=FALSE) +
jtt 11
scale_colour_brewer(palette="Set2") +
facet_wrap(~day, nrow=2) + theme(legend.position="bottom")
## Les Miserables example
data(lesmis)
lesmisnet <- merge(lesmis$edges, lesmis$vertices, by.x="from", by.y="label", all=TRUE)
p <- ggplot(data=lesmisnet, aes(from_id=from, to_id=to))
p + geom_net(layout="fruchtermanreingold")
p + geom_net(layout="fruchtermanreingold", label=TRUE, vjust=-0.5)
p + geom_net(layout="fruchtermanreingold", label=TRUE, vjust=-0.5, aes(linewidth=degree/5))
## College Football Games in the Fall 2000 regular season
# Hello world!
# Source: http://www-personal.umich.edu/~mejn/netdata/
data(football)
ftnet <- merge(football$edges, football$vertices, by.x="from", by.y="label", all=TRUE)
p <- ggplot(data=ftnet, aes(from_id=from, to_id=to))
p + geom_net(aes(colour=value), linewidth=0.75, size=4.5, ecolour="grey80") +
scale_colour_brewer("Conference", palette="Paired") + theme_net() +
theme(legend.position="bottom")
## End(Not run)

geomnet
library(ggplot2)
data(mpg)
ggplot(mpg, aes(displ, hwy)) + geom_circle(radius=0.1) + geom_point()
ggplot(mpg, aes(displ, hwy)) + geom_circle(linetype=2, radius=0.05, alpha=0.5)
ggplot(mpg, aes(displ, hwy)) + geom_circle(aes(linetype=factor(cyl)), radius=0.05, alpha=0.5)

geomnet
> library(geomnet)
> # data step: merge vertices and edges
> ftnet <- merge(
+ football$edges, football$vertices,
+ by.x = "from", by.y = "label", all = TRUE
+ )
> # label independent schools
> ftnet$schools <- ifelse(ftnet$value == "Independents", ftnet$from, "")
> # create data plot
> ggplot(data = ftnet,
+ aes(from_id = from, to_id = to)) +
+ geom_net(
+ aes(
+ colour = value, group = value,
+ linetype = factor(same.conf != 1),
+ label = schools
+ ),
+ linewidth = 0.5,
+ size = 5, vjust = -0.75, alpha = 0.3,
+ layout = 'fruchtermanreingold'
+ ) +
+ theme_net() +
+ theme(legend.position = "bottom") +
+ scale_colour_brewer("Conference", palette = "Paired") +
+ guides(linetype = FALSE)

ggstance
ibrary("ggstance")
> ggplot(mpg, aes(hwy, class, fill = factor(cyl))) +
+ geom_boxploth()
>
-------------------------------------------------------
library("ggplot2")
# Vertical
ggplot(mpg, aes(class, hwy, fill = factor(cyl))) +
geom_boxplot()
# Horizontal with coord_flip()
ggplot(mpg, aes(class, hwy, fill = factor(cyl))) +
geom_boxplot() +
coord_flip()
-----------------------------------

ggradar
library(ggradar)
suppressPackageStartupMessages(library(dplyr))
> library(scales)
> mtcars %>%
+ add_rownames( var = "group" ) %>%
+ mutate_each(funs(rescale), -group) %>%
+ tail(4) %>% select(1:10) -> mtcars_radar
> ggradar(mtcars_radar

ggsci
library("ggplot2")
data("diamonds")
ggplot(subset(diamonds, carat >= 2.2),
aes(x = table, y = price, colour = cut)) +
geom_point(alpha = 0.7) +
geom_smooth(alpha = 0.1, size = 1, span = 1) +
theme_bw() + scale_color_rickandmorty()
ggplot(subset(diamonds, carat > 2.2 & depth > 55 & depth < 70),
aes(x = depth, fill = cut)) +
geom_histogram(colour = "black", binwidth = 1, position = "dodge") +
theme_bw() + scale_fill_rickandmorty()

ggtech
library(ggtech)
>
> d <- qplot(carat, data = diamonds[diamonds$color %in%LETTERS[4:7], ], geom = "histogram", bins=30, fill = color)
> d + theme_tech(theme="airbnb") +
+ scale_fill_tech(theme="airbnb") +
+ labs(title="Airbnb theme",
+ subtitle="now with subtitles for ggplot2 >= 2.1.0")

ggnetwork
data(emon)
ggplot(emon[[1]], aes(x = x, y = y, xend = xend, yend = yend)) +
+ geom_edges(arrow = arrow(length = unit(6, "pt"), type = "closed")) +
+ geom_nodes(color = "tomato", size = 4) +
+ theme_blank()
> ggplot(ggnetwork(emon[[1]], arrow.gap = 0.04, by = "Frequency"),
+ aes(x = x, y = y, xend = xend, yend = yend)) +
+ geom_edges(arrow = arrow(length = unit(6, "pt"), type = "closed"),
+ aes(color = Sponsorship)) +
+ geom_nodes(aes(color = Sponsorship), size = 4) +
+ facet_wrap(~ Frequency) +
+ theme_facet()
ggnetwork
data(emon)
ggplot(emon[[1]], aes(x = x, y = y, xend = xend, yend = yend)) +
geom_edges(arrow = arrow(length = unit(6, "pt"), type = "closed")) +
geom_nodes(color = "tomato", size = 4) +
theme_blank()
ggthemes package
p2 <- ggplot(mtcars, aes(x = wt, y = mpg, colour = factor(gear))) +
+ geom_point() +
+ ggtitle("Cars")
p2 + theme_solarized(light = FALSE) +
scale_colour_solarized("red")
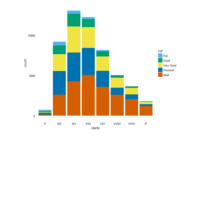
ggthemes package
ggplot(diamonds, aes(x = clarity, fill = cut)) +
geom_bar() +
theme_pander() +
scale_fill_pander()

rbokeh example
library(rbokeh)
url <- c("http://bokeh.pydata.org/en/latest/_static/images/logo.png",
"http://developer.r-project.org/Logo/Rlogo-4.png")
ss <- seq(0, 2*pi, length = 13)[-1]
ws <- runif(12, 2.5, 5) * rep(c(1, 0.8), 6)
imgdat <- data.frame(
x = sin(ss) * 10, y = cos(ss) * 10,
w = ws, h = ws * rep(c(1, 0.76), 6),
url = rep(url, 6)
)
p <- figure(xlab = "x", ylab = "y") %>%
ly_image_url(x, y, w = w, h = h, image_url = url, data = imgdat,
anchor = "center") %>%
ly_lines(sin(c(ss, ss[1])) * 10, cos(c(ss, ss[1])) * 10,
width = 15, alpha = 0.1)
p
library(rbokeh)
library(rbokeh)
>
> aapl <- read.csv('table.csv')
> aapl$Date <- as.Date(aapl$Date)
> p <- figure(title = 'Apple Stock Data') %>% ly_points(Date, Volume / (10 ^ 6), data = aapl, hover = c(Date, High, Open, Close)) %>% ly_abline(v = with(aapl, Date[which.max(Volume)])) %>% y_axis(label = 'Volume in millions', number_formatter = 'numeral', format = '0.00')
> p

plotly example
library(plotly)
z <- c(
c(8.83,8.89,8.81,8.87,8.9,8.87),
c(8.89,8.94,8.85,8.94,8.96,8.92),
c(8.84,8.9,8.82,8.92,8.93,8.91),
c(8.79,8.85,8.79,8.9,8.94,8.92),
c(8.79,8.88,8.81,8.9,8.95,8.92),
c(8.8,8.82,8.78,8.91,8.94,8.92),
c(8.75,8.78,8.77,8.91,8.95,8.92),
c(8.8,8.8,8.77,8.91,8.95,8.94),
c(8.74,8.81,8.76,8.93,8.98,8.99),
c(8.89,8.99,8.92,9.1,9.13,9.11),
c(8.97,8.97,8.91,9.09,9.11,9.11),
c(9.04,9.08,9.05,9.25,9.28,9.27),
c(9,9.01,9,9.2,9.23,9.2),
c(8.99,8.99,8.98,9.18,9.2,9.19),
c(8.93,8.97,8.97,9.18,9.2,9.18)
)
dim(z) <- c(15,6)
z2 <- z + 1
z3 <- z - 1
p <- plot_ly(z=z, type="surface",showscale=FALSE) %>%
add_trace(z=z2, type="surface", showscale=FALSE, opacity=0.98) %>%
add_trace(z=z3, type="surface", showscale=FALSE, opacity=0.98)
p
plotly example
library(plotly)
kd <- with(MASS::geyser, MASS::kde2d(duration, waiting, n = 50))
with(kd, plot_ly(x = x, y = y, z = z, type = "surface"))
rbokeh example
co2dat <- data.frame(
y = co2,
x = floor(time(co2)),
m = rep(month.abb, 39))
figure() %>%
ly_lines(x, y, group = m, data = co2dat)

point_types()
point_types()

rbokeh example
wa_cancer <- droplevels(subset(latticeExtra::USCancerRates, state == "Washington"))
## y axis sorted by male rate
ylim <- levels(with(wa_cancer, reorder(county, rate.male)))
figure(ylim = ylim, width = 700, height = 600, tools = "") %>%
ly_segments(LCL95.male, county, UCL95.male,
county, data = wa_cancer, color = NULL, width = 2) %>%
ly_points(rate.male, county, glyph = 16, data = wa_cancer)

rbokeh example
figure(ylab = "Height (inches)", width = 600) %>%
ly_boxplot(voice.part, height, data = lattice::singer)
rbokeh example
ly_baseball <- function(x) {
base_x <- c(90 * cos(pi/4), 0, 90 * cos(3 * pi/4), 0)
base_y <- c(90 * cos(pi/4), sqrt(90^2 + 90^2), 90 * sin(pi/4), 0)
distarc_x <- lapply(c(2:4) * 100, function(a)
seq(a * cos(3 * pi/4), a * cos(pi/4), length = 200))
distarc_y <- lapply(distarc_x, function(x)
sqrt((x[1]/cos(3 * pi/4))^2 - x^2))
x %>%
## boundary
ly_segments(c(0, 0), c(0, 0), c(-300, 300), c(300, 300), alpha = 0.4) %>%
## bases
ly_crect(base_x, base_y, width = 10, height = 10,
angle = 45*pi/180, color = "black", alpha = 0.4) %>%
## infield/outfield boundary
ly_curve(60.5 + sqrt(95^2 - x^2),
from = base_x[3] - 26, to = base_x[1] + 26, alpha = 0.4) %>%
## distance arcs (ly_arc should work here and would be much simpler but doesn't)
ly_multi_line(distarc_x, distarc_y, alpha = 0.4)
}
figure(xgrid = FALSE, ygrid = FALSE, width = 630, height = 540,
xlab = "Horizontal distance from home plate (ft.)",
ylab = "Vertical distance from home plate (ft.)") %>%
ly_baseball() %>%
ly_hexbin(doubles, xbins = 50, shape = 0.77, alpha = 0.75, palette = "Spectral10")
rbokeh example
figure() %>% ly_hexbin(rnorm(10000), rnorm(10000))

rbokeh example
tools <- c("pan", "wheel_zoom", "box_zoom", "box_select", "reset")
nms <- expand.grid(names(iris)[1:4], rev(names(iris)[1:4]), stringsAsFactors = FALSE)
splom_list <- vector("list", 16)
for(ii in seq_len(nrow(nms))) {
splom_list[[ii]] <- figure(width = 200, height = 200, tools = tools,
xlab = nms$Var1[ii], ylab = nms$Var2[ii]) %>%
ly_points(nms$Var1[ii], nms$Var2[ii], data = iris,
color = Species, size = 5, legend = FALSE)
}
grid_plot(splom_list, ncol = 4, same_axes = TRUE, link_data = TRUE)

rbokeh example
> library(rbokeh)
> p <- figure(width = 800, height = 400) %>%
+ ly_lines(date, Freq, data = flightfreq, alpha = 0.3) %>%
+ ly_points(date, Freq, data = flightfreq,
+ hover = list(date, Freq, dow), size = 5) %>%
+ ly_abline(v = as.Date("2001-09-11"))
> p

rbokeh
> library(maps)
> data(world.cities)
> caps <- subset(world.cities, capital == 1)
> caps$population <- prettyNum(caps$pop, big.mark = ",")
> figure(width = 800, padding_factor = 0) %>%
+ ly_map("world", col = "gray") %>%
+ ly_points(long, lat, data = caps, size = 5,
+ hover = c(name, country.etc, population))
plotly example
>library(dplyr)
> library(plotly)
> p <- ggplot2::mpg %>% group_by(class) %>%
+ summarise(mn = mean(hwy), sd = 1.96 * sd(hwy)) %>%
+ arrange(desc(mn)) %>%
+ plot_ly(x = class, y = mn, error_y = list(array = sd),
+ mode = "markers", name = "Highway") %>%
+ layout(yaxis = list(title = "Miles Per Gallon"))
> p
rbokeh
> diamonds <- ggplot2:: diamonds
> l <- levels(diamonds$color)
> plot_list <- vector(mode = 'list', 7)
>
> for (i in 1:length(l)) {
+ data <- subset(diamonds, color == l[i])
+ plot_list[[i]] <- figure(width = 350, height = 350) %>%
+ ly_points(carat, price, data = data, legend = l[i], hover = c(cut, clarity))
+ }
>
> grid_plot(plot_list, nrow = 2)

rbokeh perioidic table
library(rbokeh)
> elements <- subset(elements, !is.na(group))
> elements$group <- as.character(elements$group)
> elements$period <- as.character(elements$period)
>
> # add colors for groups
> metals <- c("alkali metal", "alkaline earth metal", "halogen",
+ "metal", "metalloid", "noble gas", "nonmetal", "transition metal")
> colors <- c("#a6cee3", "#1f78b4", "#fdbf6f", "#b2df8a", "#33a02c",
+ "#bbbb88", "#baa2a6", "#e08e79")
> elements$color <- colors[match(elements$metal, metals)]
> elements$type <- elements$metal
>
> # make coordinates for labels
> elements$symx <- paste(elements$group, ":0.1", sep = "")
> elements$numbery <- paste(elements$period, ":0.8", sep = "")
> elements$massy <- paste(elements$period, ":0.15", sep = "")
> elements$namey <- paste(elements$period, ":0.3", sep = "")
>
> # create figure
> p <- figure(title = "Periodic Table", tools = c("resize", "hover"),
+ ylim = as.character(c(7:1)), xlim = as.character(1:18),
+ xgrid = FALSE, ygrid = FALSE, xlab = "", ylab = "",
+ height = 445, width = 800) %>%
+
+ # plot rectangles
+ ly_crect(group, period, data = elements, 0.9, 0.9,
+ fill_color = color, line_color = color, fill_alpha = 0.6,
+ hover = list(name, atomic.number, type, atomic.mass,
+ electronic.configuration)) %>%
+
+ # add symbol text
+ ly_text(symx, period, text = symbol, data = elements,
+ font_style = "bold", font_size = "10pt",
+ align = "left", baseline = "middle") %>%
+
+ # add atomic number text
+ ly_text(symx, numbery, text = atomic.number, data = elements,
+ font_size = "6pt", align = "left", baseline = "middle") %>%
+
+ # add name text
+ ly_text(symx, namey, text = name, data = elements,
+ font_size = "4pt", align = "left", baseline = "middle") %>%
+
+ # add atomic mass text
+ ly_text(symx, massy, text = atomic.mass, data = elements,
+ font_size = "4pt", align = "left", baseline = "middle")
>
> p
library(rbokeh)
> library(rbokeh)
> clusters <- hclust(dist(iris[, 3:4]), method = 'average')
> clusterCut <- cutree(clusters, 3)
> p <- figure(title = 'Hierarchical Clustering of Iris Data') %>%
+ ly_points(Petal.Length, Petal.Width, data = iris, color = Species, hover = c(Sepal.Length, Sepal.Width)) %>%
+ ly_points(iris$Petal.Length, iris$Petal.Width, glyph = clusterCut, size = 13)
> p

ggraph
irisDen <- as.dendrogram(hclust(dist(iris[1:4], method='euclidean'),
method='ward.D2'))
## Add the species information to the leafs
irisDen <- dendrapply(irisDen, function(d) {
if(is.leaf(d))
attr(d, 'nodePar') <- list(species=iris[as.integer(attr(d, 'label')),5])
d
})
# Plotting this looks very much like ggplot2 except for the new geoms
ggraph(graph = irisDen, layout = 'dendrogram', repel = TRUE, circular = TRUE,
ratio = 0.5) +
geom_edge_elbow() +
geom_node_text(aes(x = x*1.05, y=y*1.05, filter=leaf,
angle = nAngle(x, y), label = label),
size=3, hjust='outward') +
geom_node_point(aes(filter=leaf, color=species)) +
coord_fixed() +
ggforce::theme_no_axes()
ggraph
>library(igraph)
>library(ggforce)
> library(ggraph)
> friendGraph <- graph_from_data_frame(highschool)
> V(friendGraph)$degree <- degree(friendGraph, mode = 'in')
> graph1957 <- subgraph.edges(friendGraph, which(E(friendGraph)$year ==1957), F)
> graph1958 <- subgraph.edges(friendGraph, which(E(friendGraph)$year ==1958), F)
> V(friendGraph)$pop.increase <- degree(graph1958, mode = 'in') >
+ degree(graph1957, mode = 'in')
>
> ggraph(friendGraph, 'igraph', algorithm = 'kk') +
+ geom_edge_fan(aes(alpha = ..index..)) +
+ geom_node_point(aes(size = degree, colour = pop.increase)) +
+ scale_edge_alpha('Friends with', guide = 'edge_direction') +
+ scale_colour_manual('Improved', values = c('firebrick', 'forestgreen')) +
+ scale_size('# Friends') +
+ facet_wrap(~year) +
+ ggforce::theme_no_axes()
>

GERGM
ibrary(GERGM)
########################### 1. No Covariates #############################
# Preparing an unbounded network without covariates for gergm estimation #
set.seed(12345)
net <- matrix(rnorm(100,0,20),10,10)
colnames(net) <- rownames(net) <- letters[1:10]
formula <- net ~ edges(method = "endogenous") + mutual + ttriads
test <- gergm(formula,
normalization_type = "division",
network_is_directed = TRUE,
number_of_networks_to_simulate = 40000,
thin = 1/10,
proposal_variance = 0.2,
MCMC_burnin = 10000,
seed = 456,
convergence_tolerance = 0.01,
force_x_theta_update = 4)
########################### 2. Covariates #############################
# Preparing an unbounded network with covariates for gergm estimation #
set.seed(12345)
net <- matrix(runif(100,0,1),10,10)
colnames(net) <- rownames(net) <- letters[1:10]
node_level_covariates <- data.frame(Age = c(25,30,34,27,36,39,27,28,35,40),
Height = c(70,70,67,58,65,67,64,74,76,80),
Type = c("A","B","B","A","A","A","B","B","C","C"))
rownames(node_level_covariates) <- letters[1:10]
network_covariate <- net + matrix(rnorm(100,0,.5),10,10)
formula <- net ~ edges(method = "regression") + mutual + ttriads + sender("Age") +
netcov("network_covariate") + nodemix("Type",base = "A")
test <- gergm(formula,
covariate_data = node_level_covariates,
number_of_networks_to_simulate = 100000,
thin = 1/10,
proposal_variance = 0.2,
MCMC_burnin = 50000,
seed = 456,
convergence_tolerance = 0.01,
force_x_theta_update = 2)
# Generate Estimate Plot
Estimate_Plot(test)
# Generate GOF Plot
GOF(test)
# Generate Trace Plot
Trace_Plot(test)
# Generate Hysteresis plots for all structural parameter estimates
hysteresis_results <- hysteresis(test,
networks_to_simulate = 1000,
burnin = 500,
range = 2,
steps = 20,
simulation_method = "Metropolis",
proposal_variance = 0.2)
GERGM
> set.seed(12345)
> net <- matrix(runif(100,0,1),10,10)
> colnames(net) <- rownames(net) <- letters[1:10]
> node_level_covariates <- data.frame(Age = c(25,30,34,27,36,39,27,28,35,40),
+ Height = c(70,70,67,58,65,67,64,74,76,80),
+ Type = c("A","B","B","A","A","A","B","B","C","C"))
> rownames(node_level_covariates) <- letters[1:10]
> network_covariate <- net + matrix(rnorm(100,0,.5),10,10)
> formula <- net ~ edges(method = "regression") + mutual + ttriads + sender("Age") +
+ netcov("network_covariate") + nodemix("Type",base = "A")
>
> test <- gergm(formula,
+ covariate_data = node_level_covariates,
+ number_of_networks_to_simulate = 100000,
+ thin = 1/10,
+ proposal_variance = 0.2,
+ MCMC_burnin = 50000,
+ seed = 456,
+ convergence_tolerance = 0.01,
+ force_x_theta_update = 2)
library(ggfortify)
library(cluster)
> autoplot(clara(iris[-5], 3))
> autoplot(fanny(iris[-5], 3), frame = TRUE)
>
library(ggfortify)
set.seed(1)
> autoplot(kmeans(USArrests, 3), data = USArrests)
> autoplot(kmeans(USArrests, 3), data = USArrests, label = TRUE, label.size = 3)
library(ggfortify)
res <- lapply(c(3, 4, 5), function(x) kmeans(iris[-5], x))
autoplot(res, data = iris[-5], ncol = 3)

library(ggfortify)
library(ggfortify)
> res <- lm(Volume ~ Girth, data = trees)
> mp <- autoplot(res, ncol = 4)
> mp

ggtern
library(ggtern)
> data(Feldspar)
> data(FeldsparRaster)
> ggtern(Feldspar,aes(Ab,An,Or)) +
+ theme_rgbw() +
+ annotation_raster_tern(FeldsparRaster,xmin=0,xmax=1,ymin=0,ymax=1) +
+ geom_mask() +
+ geom_point(size=5,aes(shape=Feldspar,fill=Feldspar),color='black') +
+ scale_shape_manual(values=c(21,24)) +
+ labs(title="Demonstration of Raster Annotation")
>

library(ggalt) library(ggplot2)
> world <- map_data("world")
> #>
> #> # maps v3.1: updated 'world': all lakes moved to separate new #
> #> # 'lakes' database. Type '?world' or 'news(package="maps")'. #
> world <- world[world$region != "Antarctica",]
>
> gg <- ggplot()
> gg <- gg + geom_map(data=world, map=world,
+ aes(x=long, y=lat, map_id=region))
> gg <- gg + coord_proj("+proj=wintri")
> gg
library(ggalt)
library(ggplot2)
> library(gridExtra)
> library(ggalt)
>
> # current verison
> packageVersion("ggalt")
[1] ‘0.1.1’
> #> [1] '0.3.0.9000'
>
> set.seed(1492)
> dat <- data.frame(x=c(1:10, 1:10, 1:10),
+ y=c(sample(15:30, 10), 2*sample(15:30, 10), 3*sample(15:30, 10)),
+ group=factor(c(rep(1, 10), rep(2, 10), rep(3, 10))))
> ggplot(dat, aes(x, y, group=group, color=group)) +
+ geom_point() +
+ geom_line()
> m <- ggplot(faithful, aes(x = eruptions, y = waiting)) +
+ geom_point() +
+ xlim(0.5, 6) +
+ ylim(40, 110)
>
> m + geom_bkde2d(bandwidth=c(0.5, 4))
> m + stat_bkde2d(bandwidth=c(0.5, 4), aes(fill = ..level..), geom = "polygon")
>

library(ggalt)
library(ggplot2)
> library(gridExtra)
> library(ggalt)
>
> # current verison
> packageVersion("ggalt")
[1] ‘0.1.1’
> #> [1] '0.3.0.9000'
>
> set.seed(1492)
> dat <- data.frame(x=c(1:10, 1:10, 1:10),
+ y=c(sample(15:30, 10), 2*sample(15:30, 10), 3*sample(15:30, 10)),
+ group=factor(c(rep(1, 10), rep(2, 10), rep(3, 10))))
> ggplot(dat, aes(x, y, group=group, color=group)) +
+ geom_point() +
+ geom_line()
> m <- ggplot(faithful, aes(x = eruptions, y = waiting)) +
+ geom_point() +
+ xlim(0.5, 6) +
+ ylim(40, 110)
>
> m + geom_bkde2d(bandwidth=c(0.5, 4))
>
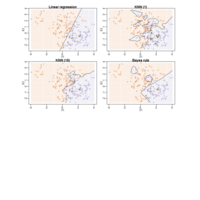
Practical Machine Learning
http://www.cbcb.umd.edu/~hcorrada/PracticalML/src/classification.R
http://www.cbcb.umd.edu/~hcorrada/PracticalML/

palette
palette(rainbow(10)); palette() palette(rainbow(10)); palette() > barplot(rep(1,20), yaxt="n", col=palette()); > palette(gray(1:10/10)); palette()0FF" "#FF0099" > barplot(rep(1,20), yaxt="n", col=palette()); > palette(gray(1:10/10)); palette()

plot mathematical expressions.
plot(1:10,type="n",xlab="",ylab="",main="plot math&numbers")
> theta<-1.23;mtext(bquote(hat(theta)==.(theta)),line=.25)
> for(i in 2:9)
+ text(i,i+1,substitute(list(xi,eta)==group("(",list(x,y),")"),list(x=i, y=i+1)))
> text(1,10, "Derivatives:"adj=0)
text(8,5, expression(paste(frac(1,sigma*sqrt(2*pi)),"",plain(e)^{frac(-(x-mu)^2, 2*sigma^2)})),cex=1.2)
ggmap-İstanbul
library(ggmap)
qmap(location = "Istanbul", zoom = 14)

rpart-Decision Tree.
library(rpart)
# grow tree
fit <- rpart(Kyphosis ~ Age + Number + Start,
method="class", data=kyphosis)
printcp(fit) # display the results
plotcp(fit) # visualize cross-validation results
summary(fit) # detailed summary of splits
# plot tree
plot(fit, uniform=TRUE,
main="Classification Tree for Kyphosis")
text(fit, use.n=TRUE, all=TRUE, cex=.8)
# create attractive postscript plot of tree
post(fit, file = "c:/tree.ps",
title = "Classification Tree for Kyphosis")
library(checkpoint) library(leaflet); library(magrittr)
## Plot all Starbucks locations using OpenStreetMap
## Credit: http://www.computerworld.com/article/2893271/business-intelligence/5-data-visualizations-in-5-minutes-each-in-5-lines-or-less-of-r.html
library(checkpoint)
checkpoint("2016-08-22")
file<- "https://opendata.socrata.com/api/views/ddym-zvjk/rows.csv"
starbucks <- read.csv(file)
library(leaflet); library(magrittr)
leaflet() %>% addTiles() %>% setView(-84.3847, 33.7613, zoom = 16) %>%
addMarkers(data = starbucks, lat = ~ Latitude, lng = ~ Longitude, popup = starbucks$Name)

Publish Plot
library(rworldmap)
> mapCountryData(mapRegion = "Turkey")

library(rworldmap)
> library(rworldmap)
> newmap <- getMap(resolution = "coarse") # different resolutions available
> plot(newmap)

dismo
mymap <- gmap("Turkey", type = "satellite")
plot(mymap)

library(dismo)
> library(sp) # classes for spatial data
> library(raster) # grids, rasters
> library(rasterVis) # raster visualisation
> library(maptools)
> library(rgeos)
> # and their dependencies
>
> library(dismo)
>
> mymap <- gmap("Turkey") # choose whatever country
> plot(mymap)
>
rWBclimate package
country.list <- c("USA", "MEX", "CAN", "BLZ")
country.dat <- get_historical_temp(country.list, "year")
ggplot(country.dat, aes(x = year, y = data, group = locator)) + geom_point() +
geom_path() + ylab("Average annual temperature of Canada") + theme_bw() +
xlab("Year") + stat_smooth(se = F, colour = "black") + facet_wrap(~locator,
scale = "free")

rWBclimate package
country.list <- c("ISL", "FIN", "NOR", "SWE")
country.dat <- get_ensemble_stats(country.list, "mavg", "tmin_means")
####### Subset data Exclude A2 scenario
country.dat.b1 <- subset(country.dat, country.dat$scenario == "b1")
# choose just one percentile
country.dat.b1 <- subset(country.dat.b1, country.dat.b1$percentile == 50)
# get just one year period
country.dat.b1 <- subset(country.dat.b1, country.dat.b1$fromYear == 2081)
ggplot(country.dat.b1, aes(x = month, y = data, group = locator, colour = locator)) +
geom_point() + geom_path() + ylab("Average daily minimum temperature") +
theme_bw() + xlab("Month")

rWBclimate package
idn.dat <- get_ensemble_precip("IDN", "mavg", 2080, 2100)
# Set line types
ltype <- rep(1, dim(idn.dat)[1])
ltype[idn.dat$percentile != 50] <- 2
idn.dat$ltype <- ltype
# Create uniqueIDs
idn.dat$uid <- paste(idn.dat$scenario, idn.dat$percentile, sep = "-")
ggplot(idn.dat, aes(x = as.factor(month), y = data, group = uid, colour = scenario,
linetype = as.factor(ltype))) + geom_point() + geom_path() + xlab("Month") +
ylab("Rain in mm") + theme_bw()

rWBclimate package
ggplot(usa.dat[usa.dat$scenario == "a2", ], aes(x = month, y = data, group = gcm,
colour = gcm)) + geom_point() + geom_path() + ylab("Average temperature in degrees C \n between 2080 and 2100") +
xlab("Month") + theme_bw()

rWBclimate package
usa.dat <- get_model_temp("USA", "mavg", 2080, 2100)
usa.dat.bcc <- usa.dat[usa.dat$gcm == "bccr_bcm2_0", ]
usa.dat.had <- usa.dat[usa.dat$gcm == "ukmo_hadcm3", ]
## Add a unique ID to each for easier plotting
usa.dat.bcc$ID <- paste(usa.dat.bcc$scenario, usa.dat.bcc$gcm, sep = "-")
usa.dat.had$ID <- paste(usa.dat.had$scenario, usa.dat.had$gcm, sep = "-")
plot.df <- rbind(usa.dat.bcc, usa.dat.had)
ggplot(plot.df, aes(x = as.factor(month), y = data, group = ID, colour = gcm,
linetype = scenario)) + geom_point() + geom_path() + ylab("Average temperature in degrees C \n between 2080 and 2100") +
xlab("Month") + theme_bw()
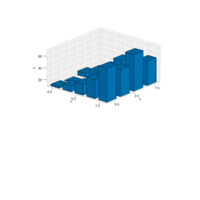
hist3D and ribbon3D
data(VADeaths)
> # hist3D and ribbon3D with greyish background, rotated, rescaled,...
> hist3D(z = VADeaths, scale = FALSE, expand = 0.01, bty = "g", phi = 20,
+ col = "#0072B2", border = "black", shade = 0.2, ltheta = 90,
+ space = 0.3, ticktype = "detailed", d = 2)
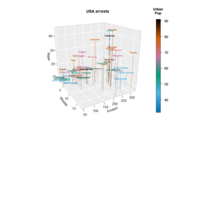
Plot3D
ref:http://www.sthda.com/english/wiki/impressive-package-for-3d-and-4d-graph-r-software-and-data-visualization

library(Amelia)- dataset:Titanic
library(Amelia)
missmap(training.data.raw, main = "Missing values vs observed")
ddpcr: Analysis and visualization of Droplet Digital PCR data in R and on the web
> plate_pnpp <- new_plate(dir, type = plate_types$fam_positive_pnpp)
Reading data files into plate... Parsed with column specification:
cols(
`Assay1 Amplitude` = col_double(),
`Assay2 Amplitude` = col_double(),
Cluster = col_integer()
)
Parsed with column specification:
cols(
`Assay1 Amplitude` = col_double(),
`Assay2 Amplitude` = col_double(),
Cluster = col_integer()
)
Parsed with column specification:
cols(
`Assay1 Amplitude` = col_double(),
`Assay2 Amplitude` = col_double(),
Cluster = col_integer()
)
Parsed with column specification:
cols(
`Assay1 Amplitude` = col_double(),
`Assay2 Amplitude` = col_double(),
Cluster = col_integer()
)
Parsed with column specification:
cols(
`Assay1 Amplitude` = col_double(),
`Assay2 Amplitude` = col_double(),
Cluster = col_integer()
)
DONE (0 seconds)
Initializing plate of type `fam_positive_pnpp`... DONE (0 seconds)
> clusters(plate_pnpp)
[1] "UNDEFINED" "FAILED" "OUTLIER" "EMPTY" "RAIN" "POSITIVE" "NEGATIVE"
> plate_pnpp <- analyze(plate_pnpp)
Identifying failed wells... DONE (0 seconds)
Identifying outlier droplets... DONE (0 seconds)
Identifying empty droplets... DONE (1 seconds)
Classifying droplets... DONE (1 seconds)
Reclassifying droplets... skipped (not enough wells with significant mutant clusters)
Analysis complete
> plate_pnpp %>% plate_meta(only_used = TRUE)
well sample row col used drops success drops_outlier drops_empty drops_non_empty
1 A01 Dean A 1 TRUE 15820 TRUE 2 13690 2130
2 A05 Dave A 5 TRUE 13165 TRUE 1 11283 1882
3 C01 Mike C 1 TRUE 14256 TRUE 0 12879 1377
4 F05 Mary F 5 TRUE 15377 TRUE 0 14126 1251
5 C05 Emily C 5 TRUE 14109 FALSE 0 NA NA
drops_empty_fraction concentration mutant_border filled_border
1 0.865 170 4194 8286
2 0.857 181 3789 8136
3 0.903 120 4356 8445
4 0.919 99 3926 8294
5 NA NA NA NA
significant_mutant_cluster mutant_num wildtype_num mutant_freq
1 FALSE 4 1827 0.218
2 TRUE 368 1224 23.100
3 FALSE 3 1248 0.240
4 TRUE 211 855 19.800
5 NA NA NA NA
> plate_pnpp %>% plot(text_size_mutant_freq = 8)

ddpcr: Analysis and visualization of Droplet Digital PCR data in R and on the web
> plate %>% plate_meta(only_used = TRUE)
well sample row col used drops
1 A01 Dean A 1 TRUE 15820
2 A05 Dave A 5 TRUE 13165
3 C01 Mike C 1 TRUE 14256
4 C05 Emily C 5 TRUE 14109
5 F05 Mary F 5 TRUE 15377
> plate <- plate %>% subset("A01:C05")
> # could have also used subset("A01, A05, C01, C05")
> plate %>% wells_used()
[1] "A01" "A05" "C01" "C05"
> plate %>% plate_data()
# A tibble: 57,350 x 4
well HEX FAM cluster
<chr> <int> <int> <int>
1 A01 577 494 1
2 A01 515 495 1
3 A01 690 645 1
4 A01 929 860 1
5 A01 844 868 1
6 A01 942 907 1
7 A01 985 923 1
8 A01 1058 966 1
9 A01 1058 979 1
10 A01 1095 1002 1
# ... with 57,340 more rows
> plate %>% plate_meta(only_used = TRUE)
well sample row col used drops
1 A01 Dean A 1 TRUE 15820
2 A05 Dave A 5 TRUE 13165
3 C01 Mike C 1 TRUE 14256
4 C05 Emily C 5 TRUE 14109
> well_info(plate, "A05", "drops_empty")
NULL
> plate %>% plot()
> well_info(plate, "A05", "drops_empty")
NULL
> plate %>% plot(wells = "A01,A05", show_full_plate = TRUE,
+ show_drops_empty = TRUE, col_drops_empty = "red",
+ title = "Show full plate")
> plate %>% plot(wells = "A01,A05", superimpose = TRUE,
+ show_grid = TRUE, show_grid_labels = TRUE, title = "Superimpose")
library(ddpcr)
library(ddpcr)
dir <- sample_data_dir()
plate <- new_plate(dir)
plot(plate)

library(ddpcr)
library(ddpcr)
dir <- sample_data_dir()
# example 1: manually set thresholds
plate1 <-
new_plate(dir, type = plate_types$custom_thresholds) %>%
subset("A01,A05") %>%
set_thresholds(c(5000, 7500)) %>%
analyze()
plot(plate1, show_grid_labels = TRUE, alpha_drops = 0.3,
title = "Manually set gating thresholds\nworks with any data")
# example 2: automatic gating
new_plate(dir, type = plate_types$fam_positive_pnpp) %>%
subset("A01:A05") %>%
analyze() %>%
plot(show_mutant_freq = FALSE, show_grid_labels = TRUE, alpha_drops = 0.3,
title = "Automatic gating\nworks with PNPP experiments")

library(ddpcr)
library(ddpcr)
dir <- sample_data_dir()
# example 1: manually set thresholds
plate1 <-
new_plate(dir, type = plate_types$custom_thresholds) %>%
subset("A01,A05") %>%
set_thresholds(c(5000, 7500)) %>%
analyze()
plot(plate1, show_grid_labels = TRUE, alpha_drops = 0.3,
title = "Manually set gating thresholds\nworks with any data")
# example 2: automatic gating
new_plate(dir, type = plate_types$fam_positive_pnpp) %>%
subset("A01:A05") %>%
analyze() %>%
plot(show_mutant_freq = FALSE, show_grid_labels = TRUE, alpha_drops = 0.3,
title = "Automatic gating\nworks with PNPP experiments")

ggmarjinal
ggMarginal(p1, type = "histogram", xparams = list(binwidth = 1, fill = "orange"))
ggmarjinal
....
ggMarginal(p1, type = "histogram")
ggmarjinal
> suppressPackageStartupMessages({
+ library("ggExtra")
+ library("ggplot2")
+ })
set.seed(30)
> df1 <- data.frame(x = rnorm(500, 50, 10), y = runif(500, 0, 50))
> (p1 <- ggplot(df1, aes(x, y)) + geom_point() + theme_bw())
> ggMarginal(p1)
> ggMarginal(p1 + theme_bw(30) + ylab("Two\nlines"))
>

plotar2
plotar(data=heart_disease, str=c('resting_blood_pressure', 'max_heart_rate'), str_target="has_heart_disease", plot_type = "histdens")

plotar
plotar(data=heart_disease, str=c('resting_blood_pressure', 'max_heart_rate'), str_target="has_heart_disease", plot_type = "histdens")
library(funModeling)
suppressMessages(library(funModeling))
data(heart_disease)
plotar(data=heart_disease, str_input="age", str_target="has_heart_disease", plot_type = "histdens")
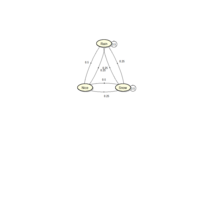
markov chains in R
library(expm)
library(markovchain)
library(diagram)
library(pracma)
stateNames <- c("Rain","Nice","Snow")
Oz <- matrix(c(.5,.25,.25,.5,0,.5,.25,.25,.5),
nrow=3, byrow=TRUE)
row.names(Oz) <- stateNames; colnames(Oz) <- stateNames
Oz
# Rain Nice Snow
# Rain 0.50 0.25 0.25
# Nice 0.50 0.00 0.50
# Snow 0.25 0.25 0.50
plotmat(Oz,pos = c(1,2),
lwd = 1, box.lwd = 2,
cex.txt = 0.8,
box.size = 0.1,
box.type = "circle",
box.prop = 0.5,
box.col = "light yellow",
arr.length=.1,
arr.width=.1,
self.cex = .4,
self.shifty = -.01,
self.shiftx = .13,
main = "")
Oz3 <- Oz %^% 3
round(Oz3,3)
# Rain Nice Snow
# Rain 0.406 0.203 0.391
# Nice 0.406 0.188 0.406
# Snow 0.391 0.203 0.406
u <- c(1/3, 1/3, 1/3)
round(u %*% Oz3,3)
#0.401 0.198 0.401

library(lattice) bwplot
> library(lattice)
> data("iris")
> bwplot(Sepal.Length~Sepal.Width, data=iris)

Turkey map
library(raster)
alt<-getData('alt',country="turkey")
plot(alt)
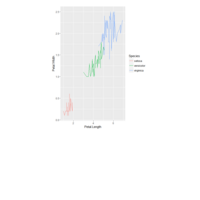
ggplot
p <- ggplot(iris, aes(Petal.Length, Petal.Width, group=Species,color=Species)) + geom_line()
p
library(plotly)
> df <- read.csv('https://cdn.rawgit.com/plotly/documentation/source/_posts/r/scattergl/weather-data.csv')
> df$Date <- zoo::as.Date(df$Date, format = "%m/%d/%Y")
> p <- plot_ly(df, x = Date, y = Mean_TemperatureC, name = "Mean Temp.", type = "scattergl", marker = list(color = "#3b3b9e")) %>% layout(title = "Mean Temparature in Seattle (1948 - 2015)", yaxis = list(title = "Temperature (<sup>o</sup>C)"))
> p

Venn Diagram
source("http://faculty.ucr.edu/~tgirke/Documents/R_BioCond/My_R_Scripts/overLapper.R") # Imports required functions.
setlist <- list(A=sample(letters, 18), B=sample(letters, 16), C=sample(letters, 20), D=sample(letters, 22), E=sample(letters, 18), F=sample(letters, 22, replace=T))
# To work with the overLapper function, the sample sets (here six) need to be stored in a list object where the different
# compontents are named by unique identifiers, here 'A to F'. These names are used as sample labels in all subsequent data
# sets and plots.
sets <- read.delim("http://faculty.ucr.edu/~tgirke/Documents/R_BioCond/Samples/sets.txt")
setlistImp <- lapply(colnames(sets), function(x) as.character(sets[sets[,x]!="", x]))
names(setlistImp) <- colnames(sets)
# Example how a list of test sets can be imported from an external table file stored in tab delimited format. Such
# a file can be easily created from a spreadsheet program, such as Excel. As a reminder, copy & paste from external
# programs into R is also possible (see read.delim function).
OLlist <- overLapper(setlist=setlist, sep="_", type="vennsets"); OLlist; names(OLlist)
# With the setting type="vennsets", the overLapper function computes all Venn Intersects for the six test samples in
# setlist and stores the results in the Venn_List component of the returned OLlist object. By default, duplicates are
# removed from the test sets. The setting keepdups=TRUE will retain duplicates by appending a counter to each entry. When
# assigning the value "intersects" to the type argument then the function will compute Regular
# Intersects instead of Venn Intersects. The Regular Intersect approach (not compatible with Venn diagrams!) is described
# in the next section. Both analyses return a present-absent matrix in the Intersect_Matrix component of OLlist. Each overlap
# set in the Venn_List data set is labeled according to the sample names provided in setlist. For instance, the composite
# name 'ABC' indicates that the entries are restricted to A, B and C. The seperator used for naming the intersect samples
# can be specified under the sep argument. By adding the argument cleanup=TRUE, one can minimize formatting issues in the
# sample sets. This setting will convert all characters in the sample sets to upper case and remove leading/trailing spaces.
#############################
## Bar plot of Venn counts ##
#############################
olBarplot(OLlist=OLlist, horiz=T, las=1, cex.names=0.6, main="Venn Bar Plot")
# Generates a bar plot for the Venn counts of the six test sample sets. In contrast to Venn diagrams, bar plots scale
# to larger numbers of sample sets. The layout of the plot can be adjusted by changing the default values of the argument:
# margins=c(4,10,3,1). The minimum number of counts to consider in the plot can be set with the mincount argument
# (default is 0). The bars themselves are colored by complexity levels using the default setting: mycol=OLlist$Complexity_Levels.
#########################
## 2-way Venn diagrams ##
#########################
setlist2 <- setlist[1:2]; OLlist2 <- overLapper(setlist=setlist2, sep="_", type="vennsets")
OLlist2$Venn_List; counts <- sapply(OLlist2$Venn_List, length); vennPlot(counts=counts)
# Plots a non-proportional 2-way Venn diagram. The main graphics features of the vennPlot function can be controlled by
# the following arguments (here with 2-way defaults): mymain="Venn Diagram": main title; mysub="default": subtitle;
# ccol=c("black","black","red"): color of counts; lcol=c("red","green"): label color; lines=c("red","green"):
# line color; mylwd=3: line width; ccex=1.0: font size of counts; lcex=1.0: font size of labels. Note: the vector
# lengths provided for the arguments ccol, lcol and lines should match the number of their corresponding features
# in the plot, e.g. 3 ccol values for a 2-way Venn diagram and 7 for a 3-way Venn diagram. The argument setlabels
# allows to provide a vector of custom sample labels. However, assigning the proper names in the original test set list
# is much more effective for tracking purposes.
#########################
## 3-way Venn diagrams ##
#########################
setlist3 <- setlist[1:3]; OLlist3 <- overLapper(setlist=setlist3, sep="_", type="vennsets")
counts <- list(sapply(OLlist3$Venn_List, length), sapply(OLlist3$Venn_List, length))
vennPlot(counts=counts, mysub="Top: var1; Bottom: var2", yoffset=c(0.3, -0.2))
# Plots a non-proportional 3-way Venn diagram. The results from several Venn comparisons can be combined in a
# single Venn diagram by assigning to the count argument a list with several count vectors. The positonal offset
# of the count sets in the plot can be controlled with the yoffset argument. The argument setting colmode=2 allows
# to assign different colors to each count set. For instance, with colmode=2 one can assign to ccol a color vector
# or a list, such as ccol=c("blue", "red") or ccol=list(1:8, 8:1).
#########################
## 4-way Venn diagrams ##
#########################
setlist4 <- setlist[1:4]
OLlist4 <- overLapper(setlist=setlist4, sep="_", type="vennsets")
counts <- list(sapply(OLlist4$Venn_List, length), sapply(OLlist4$Venn_List, length))
vennPlot(counts=counts, mysub="Top: var1; Bottom: var2", yoffset=c(0.3, -0.2))
# Plots a non-proportional 4-way Venn diagram. The setting type="circle" returns an incomplete 4-way Venn diagram as
# circles. This representation misses two overlap sectors, but is sometimes easier to navigate than the default
# ellipse version.
#########################
## 5-way Venn diagrams ##
#########################
setlist5 <- setlist[1:5]; OLlist5 <- overLapper(setlist=setlist5, sep="_", type="vennsets")
counts <- sapply(OLlist5$Venn_List, length)
vennPlot(counts=counts, ccol=c(rep(1,30),2), lcex=1.5, ccex=c(rep(1.5,5), rep(0.6,25),1.5)) # Plots a non-proportional 5-way Venn diagram.
################################
## Export and other utilities ##
################################
OLexport <- as.matrix(unlist(sapply(OLlist5[[4]], paste, collapse=" ")))
write.table(OLexport, file="test.xls", col.names=F, quote=F, sep="\t") # Exports intersect data in tabular format to a file.
OLexport <- data.frame(Venn_Comp=rep(names(OLlist5[[4]]), sapply(OLlist5[[4]], length)), IDs=unlist(OLlist5[[4]]))
write.table(OLexport, file="test.xls", row.names=F, quote=F, sep="\t") # Same as above, but exports to an alternative tabular format.
tapply(counts, OLlist5[[3]], function(x) rev(sort(x)))
# Sorts the overlap results within each complexity level by their size. This allows to identify the sample set
# combinations with the largest intersect within each complexity level.
sapply(names(setlist), function(x) table(setlist[[x]])[table(setlist[[x]])!=1])
# Command to identify and count duplicated objects in the original sample set object 'setlist'. In the given example,
# only set 'F' contains duplications. Their frequency is provided in the result.
vennPlot(counts, mymain="", mysub="", ccol="white", lcol="white") # Returns an empty Venn diagram without counts or labels.
## Typical analysis routine for sets of differentially expressed genes (DEGs)
ratio <- matrix(sample(seq(-5, 5, by=0.1), 100, replace=T), 100, 4, dimnames=list(paste("g", 1:100, sep=""), paste("DEG", 1:4, sep="")), byrow=T)
# Creates a sample matrix of gene expression log2 ratios. This could be any data type!
setlistup <- sapply(colnames(ratio), function(x) rownames(ratio[ratio[,x]>=1,]))
setlistdown <- sapply(colnames(ratio), function(x) rownames(ratio[ratio[,x]<=-1,]))
# Identifies all genes with at least a two fold up or down regulation and stores the corresponding gene identifiers
# in setlistup and setlistdown, respectively.
OLlistup <- overLapper(setlist=setlistup, sep="_", type="vennsets")
OLlistdown <- overLapper(setlist=setlistdown, sep="_", type="vennsets")
counts <- list(sapply(OLlistup$Venn_List, length), sapply(OLlistdown$Venn_List, length))
vennPlot(counts=counts, ccol=c("red", "blue"), colmode=2, mysub="Top: DEG UP; Bottom: DEG Down", yoffset=c(0.3, -0.2))
# Performs Venn analysis for the four sets stored in setlistup and setlistdown. The argument setting colmode=2 allows
# to assign different colors to each count set.
# For instance, with colmode=2 one can assign to ccol a color vector or a list, such as ccol=c("blue", "red") or ccol=list(1:8, 8:1).

library(lattice); library(gplots)
library(lattice); library(gplots)
> y <- lapply(1:4, function(x) matrix(rnorm(50), 10, 5, dimnames=list(paste("g", 1:10, sep=""), paste("t", 1:5, sep=""))))
>
> ## Plot single heatmap:
> levelplot(y[[1]])
>
> ## Arrange several heatmaps in one plot
> x1 <- levelplot(y[[1]], col.regions=colorpanel(40, "darkblue", "yellow", "white"), main="colorpanel")
> x2 <- levelplot(y[[2]], col.regions=heat.colors(75), main="heat.colors")
> x3 <- levelplot(y[[3]], col.regions=rainbow(75), main="rainbow")
> x4 <- levelplot(y[[4]], col.regions=redgreen(75), main="redgreen")
> print(x1, split=c(1,1,2,2))
> print(x2, split=c(2,1,2,2), newpage=FALSE)
> print(x3, split=c(1,2,2,2), newpage=FALSE)
> print(x4, split=c(2,2,2,2), newpage=FALSE)
library(ggplot2)
library(ggplot2)
> ggplot(data = diamonds, aes(x = carat, y = price)) + geom_point(color = "red")

lattice
library(lattice)
> xyplot(Sepal.Length ~ Sepal.Width | Species, data=iris, type="a", layout=c(1,3,1))
> parallel(~iris[1:4] | Species, iris)
> parallel(~iris[1:4] | Species, iris, horizontal.axis = FALSE, layout = c(1, 3, 1))

library(ggplot2)
library(ggplot2)
> qplot(date, uempmed, data = economics, geom = "line")

library(visNetwork) and library(igraph)
> library(visNetwork)
> library(igraph)
>
> graph.famous("Walther") %>%
+ get.data.frame( what = "both" ) %>%
+ {
+ visNetwork(
+ nodes = data.frame(
+ id = unique( c( .[["edges"]][,"from"], .[["edges"]][,"to"] ) )
+ )
+ ,edges = .[["edges"]]
+ )
+ } %>%
+ visOptions(highlightNearest = TRUE)
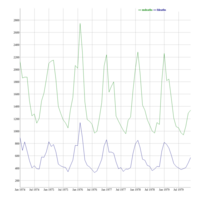
library(dygraphs)
> library(dygraphs)
> lungDeaths <- cbind(mdeaths, fdeaths)
> dygraph(lungDeaths)
library(GGally)-library(ggnet)
source:https://briatte.github.io/ggnet/

library(igraph)
library(igraph)
par(mfrow=c(2,2),mar=c(0,0,0,0), oma=c(0,0,0,0))
g = watts.strogatz.game(1,20,3,0.4)
layout.old = layout.fruchterman.reingold(g)
for(i in 1:4){
layout.new = layout.fruchterman.reingold(g,params=list(niter=10,maxdelta=2,start=layout.old))
plot(g,layout=layout.new)
layout.old = layout.new
}
networkD3
library(networkD3)
.....
diagonalNetwork(List = Flare, fontSize = 10, opacity = 0.9)

networkD3
library(networkD3)
URL <- paste0(
"https://cdn.rawgit.com/christophergandrud/networkD3/",
"master/JSONdata//flare.json")
## Convert to list format
Flare <- jsonlite::fromJSON(URL, simplifyDataFrame = FALSE)
# Use subset of data for more readable diagram
Flare$children = Flare$children[1:3]
radialNetwork(List = Flare, fontSize = 10, opacity = 0.9)

library(DiagrammeR)
Reference: https://gist.github.com/rich-iannone/de0bb88d155a2c1c7e38
Richard Ioannone
codes:
grViz("
graph severalranks {
node [shape = circle, fixedsize = true, fontcolor = '#555555',
fontname = Helvetica, fontsize = 7, style = filled,
fillcolor ='#AAAAAA', color='#555555', width = 0.12,
height = 0.12, nodesep = 0.1]
edge [color = '#AAAAAA']
graph [overlap = true, layout = neato]
node [label='', fillcolor='#000000'] n1
node [label='', fillcolor='#AAAAAA'] n2
node [label='', fillcolor='#AAAAAA'] n3
node [label='', fillcolor='#AAAAAA'] n4
node [label='', fillcolor='#AAAAAA'] n5
node [label='', fillcolor='#FFA500'] n6
node [label='', fillcolor='#FFA500'] n7
node [label='', fillcolor='#FFA500'] n8
node [label='', fillcolor='#FF00FF'] n9
node [label='', fillcolor='#555555'] n10
node [label='', fillcolor='#FFA500'] n11
node [label='', fillcolor='#FFA500'] n12
node [label='', fillcolor='#FFA500'] n13
node [label='', fillcolor='#0000FF'] n14
node [label='', fillcolor='#FFA500'] n15
node [label='', fillcolor='#FFA500'] n16
node [label='', fillcolor='#0000FF'] n17
node [label='', fillcolor='#FFA500'] n18
node [label='', fillcolor='#FFA500'] n19
node [label='', fillcolor='#0000FF'] n20
node [label='', fillcolor='#FFA500'] n21
node [label='', fillcolor='#FFA500'] n22
node [label='', fillcolor='#0000FF'] n23
node [label='', fillcolor='#FFA500'] n24
node [label='', fillcolor='#FFA500'] n25
node [label='', fillcolor='#0000FF'] n26
node [label='', fillcolor='#FFA500'] n27
node [label='', fillcolor='#FFA500'] n28
node [label='', fillcolor='#0000FF'] n29
node [label='', fillcolor='#FFA500'] n30
node [label='', fillcolor='#FFA500'] n31
node [label='', fillcolor='#0000FF'] n32
node [label='', fillcolor='#FFA500'] n33
node [label='', fillcolor='#FFA500'] n34
node [label='', fillcolor='#0000FF'] n35
node [label='', fillcolor='#FFA500'] n36
node [label='', fillcolor='#FFA500'] n37
node [label='', fillcolor='#0000FF'] n38
node [label='', fillcolor='#FFA500'] n39
node [label='', fillcolor='#FFA500'] n40
node [label='', fillcolor='#FFA500'] n41
node [label='', fillcolor='#FFA500'] n42
node [label='', fillcolor='#0000FF'] n43
node [label='', fillcolor='#FFA500'] n44
node [label='', fillcolor='#00FFFF'] n45
node [label='', fillcolor='#FFA500'] n46
node [label='', fillcolor='#FFA500'] n47
node [label='', fillcolor='#0000FF'] n48
node [label='', fillcolor='#0000FF'] n49
node [label='', fillcolor='#0000FF'] n50
node [label='', fillcolor='#0000FF'] n51
node [label='', fillcolor='#0000FF'] n52
node [label='', fillcolor='#555555'] n53
node [label='', fillcolor='#0000FF'] n54
node [label='', fillcolor='#0000FF'] n55
node [label='', fillcolor='#0000FF'] n56
node [label='', fillcolor='#0000FF'] n57
node [label='', fillcolor='#0000FF'] n58
node [label='', fillcolor='#555555'] n59
node [label='', fillcolor='#0000FF'] n60
node [label='', fillcolor='#00FFFF'] n61
node [label='', fillcolor='#0000FF'] n62
node [label='', fillcolor='#F0E68C'] n63
node [label='', fillcolor='#F0E68C'] n64
node [label='', fillcolor='#0000FF'] n65
node [label='', fillcolor='#F0E68C'] n66
node [label='', fillcolor='#0000FF'] n67
node [label='', fillcolor='#0000FF'] n68
node [label='', fillcolor='#F0E68C'] n69
node [label='', fillcolor='#0000FF'] n70
node [label='', fillcolor='#F0E68C'] n71
node [label='', fillcolor='#0000FF'] n72
node [label='', fillcolor='#F0E68C'] n73
node [label='', fillcolor='#0000FF'] n74
node [label='', fillcolor='#555555'] n75
node [label='', fillcolor='#F0E68C'] n76
node [label='', fillcolor='#0000FF'] n77
node [label='', fillcolor='#F0E68C'] n78
node [label='', fillcolor='#0000FF'] n79
node [label='', fillcolor='#F0E68C'] n80
node [label='', fillcolor='#0000FF'] n81
node [label='', fillcolor='#F0E68C'] n82
node [label='', fillcolor='#0000FF'] n83
node [label='', fillcolor='#F0E68C'] n84
node [label='', fillcolor='#0000FF'] n85
node [label='', fillcolor='#0000FF'] n86
node [label='', fillcolor='#F0E68C'] n87
node [label='', fillcolor='#0000FF'] n88
node [label='', fillcolor='#F0E68C'] n89
node [label='', fillcolor='#0000FF'] n90
node [label='', fillcolor='#0000FF'] n91
node [label='', fillcolor='#F0E68C'] n92
node [label='', fillcolor='#0000FF'] n93
node [label='', fillcolor='#0000FF'] n94
node [label='', fillcolor='#F0E68C'] n95
node [label='', fillcolor='#0000FF'] n96
node [label='', fillcolor='#0000FF'] n97
node [label='', fillcolor='#00FFFF'] n98
node [label='', fillcolor='#0000FF'] n99
node [label='', fillcolor='#F0E68C'] n100
node [label='', fillcolor='#F0E68C'] n101
node [label='', fillcolor='#0000FF'] n102
node [label='', fillcolor='#0000FF'] n103
node [label='', fillcolor='#F0E68C'] n104
node [label='', fillcolor='#0000FF'] n105
node [label='', fillcolor='#0000FF'] n106
node [label='', fillcolor='#F0E68C'] n107
node [label='', fillcolor='#0000FF'] n108
node [label='', fillcolor='#F0E68C'] n109
node [label='', fillcolor='#0000FF'] n110
node [label='', fillcolor='#F0E68C'] n111
node [label='', fillcolor='#0000FF'] n112
node [label='', fillcolor='#F0E68C'] n113
node [label='', fillcolor='#0000FF'] n114
node [label='', fillcolor='#F0E68C'] n115
node [label='', fillcolor='#0000FF'] n116
node [label='', fillcolor='#F0E68C'] n117
node [label='', fillcolor='#0000FF'] n118
node [label='', fillcolor='#F0E68C'] n119
node [label='', fillcolor='#0000FF'] n120
node [label='', fillcolor='#F0E68C'] n121
node [label='', fillcolor='#0000FF'] n122
node [label='', fillcolor='#F0E68C'] n123
node [label='', fillcolor='#0000FF'] n124
node [label='', fillcolor='#F0E68C'] n125
node [label='', fillcolor='#0000FF'] n126
node [label='', fillcolor='#F0E68C'] n127
node [label='', fillcolor='#0000FF'] n128
node [label='', fillcolor='#F0E68C'] n129
node [label='', fillcolor='#0000FF'] n130
node [label='', fillcolor='#F0E68C'] n131
node [label='', fillcolor='#0000FF'] n132
node [label='', fillcolor='#F0E68C'] n133
node [label='', fillcolor='#0000FF'] n134
node [label='', fillcolor='#F0E68C'] n135
node [label='', fillcolor='#0000FF'] n136
node [label='', fillcolor='#F0E68C'] n137
node [label='', fillcolor='#0000FF'] n138
node [label='', fillcolor='#555555'] n139
node [label='', fillcolor='#0000FF'] n140
node [label='', fillcolor='#F0E68C'] n141
node [label='', fillcolor='#0000FF'] n142
node [label='', fillcolor='#F0E68C'] n143
node [label='', fillcolor='#0000FF'] n144
node [label='', fillcolor='#F0E68C'] n145
node [label='', fillcolor='#0000FF'] n146
node [label='', fillcolor='#F0E68C'] n147
node [label='', fillcolor='#0000FF'] n148
node [label='', fillcolor='#F0E68C'] n149
node [label='', fillcolor='#0000FF'] n150
node [label='', fillcolor='#555555'] n151
node [label='', fillcolor='#0000FF'] n152
node [label='', fillcolor='#555555'] n153
node [label='', fillcolor='#0000FF'] n154
node [label='', fillcolor='#00FFFF'] n155
node [label='', fillcolor='#0000FF'] n156
node [label='', fillcolor='#F0E68C'] n157
node [label='', fillcolor='#F0E68C'] n158
node [label='', fillcolor='#0000FF'] n159
node [label='', fillcolor='#F0E68C'] n160
node [label='', fillcolor='#0000FF'] n161
node [label='', fillcolor='#F0E68C'] n162
node [label='', fillcolor='#0000FF'] n163
node [label='', fillcolor='#0000FF'] n164
node [label='', fillcolor='#F0E68C'] n165
node [label='', fillcolor='#0000FF'] n166
node [label='', fillcolor='#F0E68C'] n167
node [label='', fillcolor='#0000FF'] n168
node [label='', fillcolor='#F0E68C'] n169
node [label='', fillcolor='#0000FF'] n170
node [label='', fillcolor='#F0E68C'] n171
node [label='', fillcolor='#0000FF'] n172
node [label='', fillcolor='#F0E68C'] n173
node [label='', fillcolor='#0000FF'] n174
node [label='', fillcolor='#F0E68C'] n175
node [label='', fillcolor='#0000FF'] n176
node [label='', fillcolor='#F0E68C'] n177
node [label='', fillcolor='#0000FF'] n178
node [label='', fillcolor='#F0E68C'] n179
node [label='', fillcolor='#0000FF'] n180
node [label='', fillcolor='#0000FF'] n181
node [label='', fillcolor='#0000FF'] n182
node [label='', fillcolor='#F0E68C'] n183
node [label='', fillcolor='#0000FF'] n184
node [label='', fillcolor='#00FFFF'] n185
node [label='', fillcolor='#0000FF'] n186
node [label='', fillcolor='#F0E68C'] n187
node [label='', fillcolor='#F0E68C'] n188
node [label='', fillcolor='#0000FF'] n189
node [label='', fillcolor='#F0E68C'] n190
node [label='', fillcolor='#0000FF'] n191
node [label='', fillcolor='#0000FF'] n192
node [label='', fillcolor='#F0E68C'] n193
node [label='', fillcolor='#0000FF'] n194
node [label='', fillcolor='#F0E68C'] n195
node [label='', fillcolor='#0000FF'] n196
node [label='', fillcolor='#F0E68C'] n197
node [label='', fillcolor='#F0E68C'] n198
node [label='', fillcolor='#0000FF'] n199
node [label='', fillcolor='#F0E68C'] n200
node [label='', fillcolor='#0000FF'] n201
node [label='', fillcolor='#F0E68C'] n202
node [label='', fillcolor='#0000FF'] n203
node [label='', fillcolor='#0000FF'] n204
node [label='', fillcolor='#F0E68C'] n205
node [label='', fillcolor='#0000FF'] n206
node [label='', fillcolor='#0000FF'] n207
node [label='', fillcolor='#F0E68C'] n208
node [label='', fillcolor='#0000FF'] n209
node [label='', fillcolor='#F0E68C'] n210
node [label='', fillcolor='#0000FF'] n211
node [label='', fillcolor='#F0E68C'] n212
node [label='', fillcolor='#0000FF'] n213
node [label='', fillcolor='#F0E68C'] n214
node [label='', fillcolor='#0000FF'] n215
node [label='', fillcolor='#0000FF'] n216
node [label='', fillcolor='#F0E68C'] n217
node [label='', fillcolor='#0000FF'] n218
node [label='', fillcolor='#0000FF'] n219
node [label='', fillcolor='#F0E68C'] n220
node [label='', fillcolor='#0000FF'] n221
node [label='', fillcolor='#F0E68C'] n222
node [label='', fillcolor='#0000FF'] n223
node [label='', fillcolor='#F0E68C'] n224
node [label='', fillcolor='#0000FF'] n225
node [label='', fillcolor='#F0E68C'] n226
node [label='', fillcolor='#0000FF'] n227
node [label='', fillcolor='#F0E68C'] n228
node [label='', fillcolor='#0000FF'] n229
node [label='', fillcolor='#00FFFF'] n230
node [label='', fillcolor='#0000FF'] n231
node [label='', fillcolor='#F0E68C'] n232
node [label='', fillcolor='#F0E68C'] n233
node [label='', fillcolor='#0000FF'] n234
node [label='', fillcolor='#F0E68C'] n235
node [label='', fillcolor='#0000FF'] n236
node [label='', fillcolor='#0000FF'] n237
node [label='', fillcolor='#F0E68C'] n238
node [label='', fillcolor='#0000FF'] n239
node [label='', fillcolor='#0000FF'] n240
node [label='', fillcolor='#555555'] n241
node [label='', fillcolor='#0000FF'] n242
node [label='', fillcolor='#555555'] n243
node [label='', fillcolor='#0000FF'] n244
node [label='', fillcolor='#555555'] n245
node [label='', fillcolor='#F0E68C'] n246
node [label='', fillcolor='#0000FF'] n247
node [label='', fillcolor='#F0E68C'] n248
node [label='', fillcolor='#0000FF'] n249
node [label='', fillcolor='#00FFFF'] n250
node [label='', fillcolor='#0000FF'] n251
node [label='', fillcolor='#F0E68C'] n252
node [label='', fillcolor='#F0E68C'] n253
node [label='', fillcolor='#0000FF'] n254
node [label='', fillcolor='#F0E68C'] n255
node [label='', fillcolor='#0000FF'] n256
node [label='', fillcolor='#F0E68C'] n257
node [label='', fillcolor='#0000FF'] n258
node [label='', fillcolor='#0000FF'] n259
node [label='', fillcolor='#F0E68C'] n260
node [label='', fillcolor='#0000FF'] n261
node [label='', fillcolor='#F0E68C'] n262
node [label='', fillcolor='#0000FF'] n263
node [label='', fillcolor='#F0E68C'] n264
node [label='', fillcolor='#0000FF'] n265
node [label='', fillcolor='#F0E68C'] n266
node [label='', fillcolor='#0000FF'] n267
node [label='', fillcolor='#F0E68C'] n268
node [label='', fillcolor='#0000FF'] n269
node [label='', fillcolor='#F0E68C'] n270
node [label='', fillcolor='#0000FF'] n271
node [label='', fillcolor='#F0E68C'] n272
node [label='', fillcolor='#0000FF'] n273
node [label='', fillcolor='#F0E68C'] n274
node [label='', fillcolor='#0000FF'] n275
node [label='', fillcolor='#F0E68C'] n276
node [label='', fillcolor='#0000FF'] n277
node [label='', fillcolor='#0000FF'] n278
node [label='', fillcolor='#F0E68C'] n279
node [label='', fillcolor='#0000FF'] n280
node [label='', fillcolor='#F0E68C'] n281
node [label='', fillcolor='#0000FF'] n282
node [label='', fillcolor='#0000FF'] n283
node [label='', fillcolor='#F0E68C'] n284
node [label='', fillcolor='#0000FF'] n285
node [label='', fillcolor='#0000FF'] n286
node [label='', fillcolor='#F0E68C'] n287
node [label='', fillcolor='#0000FF'] n288
node [label='', fillcolor='#F0E68C'] n289
node [label='', fillcolor='#0000FF'] n290
node [label='', fillcolor='#F0E68C'] n291
node [label='', fillcolor='#0000FF'] n292
node [label='', fillcolor='#F0E68C'] n293
node [label='', fillcolor='#0000FF'] n294
node [label='', fillcolor='#F0E68C'] n295
node [label='', fillcolor='#0000FF'] n296
node [label='', fillcolor='#F0E68C'] n297
node [label='', fillcolor='#0000FF'] n298
node [label='', fillcolor='#F0E68C'] n299
node [label='', fillcolor='#0000FF'] n300
node [label='', fillcolor='#F0E68C'] n301
node [label='', fillcolor='#0000FF'] n302
node [label='', fillcolor='#F0E68C'] n303
node [label='', fillcolor='#0000FF'] n304
node [label='', fillcolor='#F0E68C'] n305
node [label='', fillcolor='#0000FF'] n306
node [label='', fillcolor='#F0E68C'] n307
node [label='', fillcolor='#0000FF'] n308
node [label='', fillcolor='#F0E68C'] n309
node [label='', fillcolor='#0000FF'] n310
node [label='', fillcolor='#F0E68C'] n311
node [label='', fillcolor='#0000FF'] n312
node [label='', fillcolor='#F0E68C'] n313
node [label='', fillcolor='#0000FF'] n314
node [label='', fillcolor='#F0E68C'] n315
node [label='', fillcolor='#0000FF'] n316
node [label='', fillcolor='#F0E68C'] n317
node [label='', fillcolor='#0000FF'] n318
node [label='', fillcolor='#F0E68C'] n319
node [label='', fillcolor='#0000FF'] n320
node [label='', fillcolor='#F0E68C'] n321
node [label='', fillcolor='#0000FF'] n322
node [label='', fillcolor='#F0E68C'] n323
node [label='', fillcolor='#0000FF'] n324
node [label='', fillcolor='#00FFFF'] n325
node [label='', fillcolor='#0000FF'] n326
node [label='', fillcolor='#F0E68C'] n327
node [label='', fillcolor='#F0E68C'] n328
node [label='', fillcolor='#0000FF'] n329
node [label='', fillcolor='#0000FF'] n330
node [label='', fillcolor='#F0E68C'] n331
node [label='', fillcolor='#555555'] n332
node [label='', fillcolor='#0000FF'] n333
node [label='', fillcolor='#0000FF'] n334
node [label='', fillcolor='#0000FF'] n335
node [label='', fillcolor='#F0E68C'] n336
node [label='', fillcolor='#0000FF'] n337
node [label='', fillcolor='#0000FF'] n338
node [label='', fillcolor='#F0E68C'] n339
node [label='', fillcolor='#0000FF'] n340
node [label='', fillcolor='#0000FF'] n341
node [label='', fillcolor='#0000FF'] n342
node [label='', fillcolor='#F0E68C'] n343
node [label='', fillcolor='#0000FF'] n344
node [label='', fillcolor='#555555'] n345
node [label='', fillcolor='#00FFFF'] n346
node [label='', fillcolor='#0000FF'] n347
node [label='', fillcolor='#F0E68C'] n348
node [label='', fillcolor='#F0E68C'] n349
node [label='', fillcolor='#0000FF'] n350
node [label='', fillcolor='#F0E68C'] n351
node [label='', fillcolor='#0000FF'] n352
node [label='', fillcolor='#0000FF'] n353
node [label='', fillcolor='#F0E68C'] n354
node [label='', fillcolor='#0000FF'] n355
node [label='', fillcolor='#F0E68C'] n356
node [label='', fillcolor='#0000FF'] n357
node [label='', fillcolor='#F0E68C'] n358
node [label='', fillcolor='#0000FF'] n359
node [label='', fillcolor='#F0E68C'] n360
node [label='', fillcolor='#0000FF'] n361
node [label='', fillcolor='#F0E68C'] n362
node [label='', fillcolor='#0000FF'] n363
node [label='', fillcolor='#F0E68C'] n364
node [label='', fillcolor='#0000FF'] n365
node [label='', fillcolor='#F0E68C'] n366
node [label='', fillcolor='#0000FF'] n367
node [label='', fillcolor='#00FFFF'] n368
node [label='', fillcolor='#0000FF'] n369
node [label='', fillcolor='#F0E68C'] n370
node [label='', fillcolor='#F0E68C'] n371
node [label='', fillcolor='#0000FF'] n372
node [label='', fillcolor='#F0E68C'] n373
node [label='', fillcolor='#0000FF'] n374
node [label='', fillcolor='#F0E68C'] n375
node [label='', fillcolor='#0000FF'] n376
node [label='', fillcolor='#00FFFF'] n377
node [label='', fillcolor='#0000FF'] n378
node [label='', fillcolor='#F0E68C'] n379
node [label='', fillcolor='#F0E68C'] n380
node [label='', fillcolor='#0000FF'] n381
node [label='', fillcolor='#F0E68C'] n382
node [label='', fillcolor='#0000FF'] n383
node [label='', fillcolor='#FF00FF'] n384
node [label='', fillcolor='#F0E68C'] n385
node [label='', fillcolor='#0000FF'] n386
node [label='', fillcolor='#F0E68C'] n387
node [label='', fillcolor='#0000FF'] n388
node [label='', fillcolor='#FF00FF'] n389
node [label='', fillcolor='#F0E68C'] n390
node [label='', fillcolor='#0000FF'] n391
node [label='', fillcolor='#FF00FF'] n392
node [label='', fillcolor='#F0E68C'] n393
node [label='', fillcolor='#0000FF'] n394
node [label='', fillcolor='#FF00FF'] n395
node [label='', fillcolor='#F0E68C'] n396
node [label='', fillcolor='#0000FF'] n397
node [label='', fillcolor='#FF00FF'] n398
node [label='', fillcolor='#F0E68C'] n399
node [label='', fillcolor='#0000FF'] n400
node [label='', fillcolor='#FF00FF'] n401
node [label='', fillcolor='#F0E68C'] n402
node [label='', fillcolor='#0000FF'] n403
node [label='', fillcolor='#FF00FF'] n404
node [label='', fillcolor='#F0E68C'] n405
node [label='', fillcolor='#0000FF'] n406
node [label='', fillcolor='#FF00FF'] n407
n1 -- n2 [len=0.800]
n1 -- n5 [len=0.800]
n2 -- n3 [len=0.728]
n2 -- n4 [len=0.728]
n5 -- n6 [len=0.728]
n6 -- n7 [len=0.656]
n6 -- n46 [len=0.656]
n7 -- n8 [len=0.583]
n7 -- n44 [len=0.583]
n8 -- n9 [len=0.511]
n8 -- n10 [len=0.511]
n10 -- n11 [len=0.439]
n11 -- n12 [len=0.367]
n11 -- n15 [len=0.367]
n11 -- n18 [len=0.367]
n11 -- n21 [len=0.367]
n11 -- n24 [len=0.367]
n11 -- n27 [len=0.367]
n11 -- n30 [len=0.367]
n11 -- n33 [len=0.367]
n11 -- n36 [len=0.367]
n11 -- n39 [len=0.367]
n11 -- n41 [len=0.367]
n12 -- n13 [len=0.294]
n13 -- n14 [len=0.222]
n15 -- n16 [len=0.294]
n16 -- n17 [len=0.222]
n18 -- n19 [len=0.294]
n19 -- n20 [len=0.222]
n21 -- n22 [len=0.294]
n22 -- n23 [len=0.222]
n24 -- n25 [len=0.294]
n25 -- n26 [len=0.222]
n27 -- n28 [len=0.294]
n28 -- n29 [len=0.222]
n30 -- n31 [len=0.294]
n31 -- n32 [len=0.222]
n33 -- n34 [len=0.294]
n34 -- n35 [len=0.222]
n36 -- n37 [len=0.294]
n37 -- n38 [len=0.222]
n39 -- n40 [len=0.294]
n41 -- n42 [len=0.294]
n42 -- n43 [len=0.222]
n44 -- n45 [len=0.511]
n46 -- n47 [len=0.583]
n47 -- n48 [len=0.511]
n47 -- n49 [len=0.511]
n47 -- n50 [len=0.511]
n47 -- n51 [len=0.511]
n47 -- n52 [len=0.511]
n47 -- n53 [len=0.511]
n47 -- n54 [len=0.511]
n47 -- n55 [len=0.511]
n47 -- n56 [len=0.511]
n47 -- n57 [len=0.511]
n47 -- n58 [len=0.511]
n47 -- n59 [len=0.511]
n47 -- n61 [len=0.511]
n47 -- n63 [len=0.511]
n47 -- n98 [len=0.511]
n47 -- n100 [len=0.511]
n47 -- n155 [len=0.511]
n47 -- n157 [len=0.511]
n47 -- n185 [len=0.511]
n47 -- n187 [len=0.511]
n47 -- n230 [len=0.511]
n47 -- n232 [len=0.511]
n47 -- n250 [len=0.511]
n47 -- n252 [len=0.511]
n47 -- n325 [len=0.511]
n47 -- n327 [len=0.511]
n47 -- n346 [len=0.511]
n47 -- n348 [len=0.511]
n47 -- n368 [len=0.511]
n47 -- n370 [len=0.511]
n47 -- n377 [len=0.511]
n47 -- n379 [len=0.511]
n59 -- n60 [len=0.439]
n61 -- n62 [len=0.439]
n63 -- n64 [len=0.439]
n63 -- n66 [len=0.439]
n63 -- n69 [len=0.439]
n63 -- n71 [len=0.439]
n63 -- n73 [len=0.439]
n63 -- n76 [len=0.439]
n63 -- n78 [len=0.439]
n63 -- n80 [len=0.439]
n63 -- n82 [len=0.439]
n63 -- n84 [len=0.439]
n63 -- n87 [len=0.439]
n63 -- n89 [len=0.439]
n63 -- n92 [len=0.439]
n63 -- n95 [len=0.439]
n64 -- n65 [len=0.367]
n66 -- n67 [len=0.367]
n66 -- n68 [len=0.367]
n69 -- n70 [len=0.367]
n71 -- n72 [len=0.367]
n73 -- n74 [len=0.367]
n73 -- n75 [len=0.367]
n76 -- n77 [len=0.367]
n78 -- n79 [len=0.367]
n80 -- n81 [len=0.367]
n82 -- n83 [len=0.367]
n84 -- n85 [len=0.367]
n85 -- n86 [len=0.294]
n87 -- n88 [len=0.367]
n89 -- n90 [len=0.367]
n90 -- n91 [len=0.294]
n92 -- n93 [len=0.367]
n93 -- n94 [len=0.294]
n95 -- n96 [len=0.367]
n96 -- n97 [len=0.294]
n98 -- n99 [len=0.439]
n100 -- n101 [len=0.439]
n100 -- n104 [len=0.439]
n100 -- n107 [len=0.439]
n100 -- n109 [len=0.439]
n100 -- n111 [len=0.439]
n100 -- n113 [len=0.439]
n100 -- n115 [len=0.439]
n100 -- n117 [len=0.439]
n100 -- n119 [len=0.439]
n100 -- n121 [len=0.439]
n100 -- n123 [len=0.439]
n100 -- n125 [len=0.439]
n100 -- n127 [len=0.439]
n100 -- n129 [len=0.439]
n100 -- n131 [len=0.439]
n100 -- n133 [len=0.439]
n100 -- n135 [len=0.439]
n100 -- n137 [len=0.439]
n100 -- n141 [len=0.439]
n100 -- n143 [len=0.439]
n100 -- n145 [len=0.439]
n100 -- n147 [len=0.439]
n100 -- n149 [len=0.439]
n101 -- n102 [len=0.367]
n101 -- n103 [len=0.367]
n104 -- n105 [len=0.367]
n104 -- n106 [len=0.367]
n107 -- n108 [len=0.367]
n109 -- n110 [len=0.367]
n111 -- n112 [len=0.367]
n113 -- n114 [len=0.367]
n115 -- n116 [len=0.367]
n117 -- n118 [len=0.367]
n119 -- n120 [len=0.367]
n121 -- n122 [len=0.367]
n123 -- n124 [len=0.367]
n125 -- n126 [len=0.367]
n127 -- n128 [len=0.367]
n129 -- n130 [len=0.367]
n131 -- n132 [len=0.367]
n133 -- n134 [len=0.367]
n135 -- n136 [len=0.367]
n137 -- n138 [len=0.367]
n137 -- n139 [len=0.367]
n137 -- n140 [len=0.367]
n141 -- n142 [len=0.367]
n143 -- n144 [len=0.367]
n145 -- n146 [len=0.367]
n147 -- n148 [len=0.367]
n149 -- n150 [len=0.367]
n149 -- n151 [len=0.367]
n149 -- n152 [len=0.367]
n149 -- n153 [len=0.367]
n149 -- n154 [len=0.367]
n155 -- n156 [len=0.439]
n157 -- n158 [len=0.439]
n157 -- n160 [len=0.439]
n157 -- n162 [len=0.439]
n157 -- n165 [len=0.439]
n157 -- n167 [len=0.439]
n157 -- n169 [len=0.439]
n157 -- n171 [len=0.439]
n157 -- n173 [len=0.439]
n157 -- n175 [len=0.439]
n157 -- n177 [len=0.439]
n157 -- n179 [len=0.439]
n157 -- n183 [len=0.439]
n158 -- n159 [len=0.367]
n160 -- n161 [len=0.367]
n162 -- n163 [len=0.367]
n162 -- n164 [len=0.367]
n165 -- n166 [len=0.367]
n167 -- n168 [len=0.367]
n169 -- n170 [len=0.367]
n171 -- n172 [len=0.367]
n173 -- n174 [len=0.367]
n175 -- n176 [len=0.367]
n177 -- n178 [len=0.367]
n179 -- n180 [len=0.367]
n179 -- n181 [len=0.367]
n179 -- n182 [len=0.367]
n183 -- n184 [len=0.367]
n185 -- n186 [len=0.439]
n187 -- n188 [len=0.439]
n187 -- n190 [len=0.439]
n187 -- n193 [len=0.439]
n187 -- n195 [len=0.439]
n187 -- n197 [len=0.439]
n187 -- n198 [len=0.439]
n187 -- n200 [len=0.439]
n187 -- n202 [len=0.439]
n187 -- n205 [len=0.439]
n187 -- n208 [len=0.439]
n187 -- n210 [len=0.439]
n187 -- n212 [len=0.439]
n187 -- n214 [len=0.439]
n187 -- n217 [len=0.439]
n187 -- n220 [len=0.439]
n187 -- n222 [len=0.439]
n187 -- n224 [len=0.439]
n187 -- n226 [len=0.439]
n187 -- n228 [len=0.439]
n188 -- n189 [len=0.367]
n190 -- n191 [len=0.367]
n190 -- n192 [len=0.367]
n193 -- n194 [len=0.367]
n195 -- n196 [len=0.367]
n198 -- n199 [len=0.367]
n200 -- n201 [len=0.367]
n202 -- n203 [len=0.367]
n202 -- n204 [len=0.367]
n205 -- n206 [len=0.367]
n205 -- n207 [len=0.367]
n208 -- n209 [len=0.367]
n210 -- n211 [len=0.367]
n212 -- n213 [len=0.367]
n214 -- n215 [len=0.367]
n214 -- n216 [len=0.367]
n217 -- n218 [len=0.367]
n217 -- n219 [len=0.367]
n220 -- n221 [len=0.367]
n222 -- n223 [len=0.367]
n224 -- n225 [len=0.367]
n226 -- n227 [len=0.367]
n228 -- n229 [len=0.367]
n230 -- n231 [len=0.439]
n232 -- n233 [len=0.439]
n232 -- n235 [len=0.439]
n232 -- n238 [len=0.439]
n232 -- n246 [len=0.439]
n232 -- n248 [len=0.439]
n233 -- n234 [len=0.367]
n235 -- n236 [len=0.367]
n235 -- n237 [len=0.367]
n238 -- n239 [len=0.367]
n238 -- n240 [len=0.367]
n238 -- n242 [len=0.367]
n238 -- n244 [len=0.367]
n240 -- n241 [len=0.294]
n242 -- n243 [len=0.294]
n244 -- n245 [len=0.294]
n246 -- n247 [len=0.367]
n248 -- n249 [len=0.367]
n250 -- n251 [len=0.439]
n252 -- n253 [len=0.439]
n252 -- n255 [len=0.439]
n252 -- n257 [len=0.439]
n252 -- n260 [len=0.439]
n252 -- n262 [len=0.439]
n252 -- n264 [len=0.439]
n252 -- n266 [len=0.439]
n252 -- n268 [len=0.439]
n252 -- n270 [len=0.439]
n252 -- n272 [len=0.439]
n252 -- n274 [len=0.439]
n252 -- n276 [len=0.439]
n252 -- n279 [len=0.439]
n252 -- n281 [len=0.439]
n252 -- n284 [len=0.439]
n252 -- n287 [len=0.439]
n252 -- n289 [len=0.439]
n252 -- n291 [len=0.439]
n252 -- n293 [len=0.439]
n252 -- n295 [len=0.439]
n252 -- n297 [len=0.439]
n252 -- n299 [len=0.439]
n252 -- n301 [len=0.439]
n252 -- n303 [len=0.439]
n252 -- n305 [len=0.439]
n252 -- n307 [len=0.439]
n252 -- n309 [len=0.439]
n252 -- n311 [len=0.439]
n252 -- n313 [len=0.439]
n252 -- n315 [len=0.439]
n252 -- n317 [len=0.439]
n252 -- n319 [len=0.439]
n252 -- n321 [len=0.439]
n252 -- n323 [len=0.439]
n253 -- n254 [len=0.367]
n255 -- n256 [len=0.367]
n257 -- n258 [len=0.367]
n257 -- n259 [len=0.367]
n260 -- n261 [len=0.367]
n262 -- n263 [len=0.367]
n264 -- n265 [len=0.367]
n266 -- n267 [len=0.367]
n268 -- n269 [len=0.367]
n270 -- n271 [len=0.367]
n272 -- n273 [len=0.367]
n274 -- n275 [len=0.367]
n276 -- n277 [len=0.367]
n276 -- n278 [len=0.367]
n279 -- n280 [len=0.367]
n281 -- n282 [len=0.367]
n281 -- n283 [len=0.367]
n284 -- n285 [len=0.367]
n284 -- n286 [len=0.367]
n287 -- n288 [len=0.367]
n289 -- n290 [len=0.367]
n291 -- n292 [len=0.367]
n293 -- n294 [len=0.367]
n295 -- n296 [len=0.367]
n297 -- n298 [len=0.367]
n299 -- n300 [len=0.367]
n301 -- n302 [len=0.367]
n303 -- n304 [len=0.367]
n305 -- n306 [len=0.367]
n307 -- n308 [len=0.367]
n309 -- n310 [len=0.367]
n311 -- n312 [len=0.367]
n313 -- n314 [len=0.367]
n315 -- n316 [len=0.367]
n317 -- n318 [len=0.367]
n319 -- n320 [len=0.367]
n321 -- n322 [len=0.367]
n323 -- n324 [len=0.367]
n325 -- n326 [len=0.439]
n327 -- n328 [len=0.439]
n327 -- n331 [len=0.439]
n327 -- n336 [len=0.439]
n327 -- n339 [len=0.439]
n327 -- n343 [len=0.439]
n328 -- n329 [len=0.367]
n328 -- n330 [len=0.367]
n331 -- n332 [len=0.367]
n331 -- n333 [len=0.367]
n331 -- n334 [len=0.367]
n331 -- n335 [len=0.367]
n336 -- n337 [len=0.367]
n336 -- n338 [len=0.367]
n339 -- n340 [len=0.367]
n339 -- n341 [len=0.367]
n339 -- n342 [len=0.367]
n343 -- n344 [len=0.367]
n344 -- n345 [len=0.294]
n346 -- n347 [len=0.439]
n348 -- n349 [len=0.439]
n348 -- n351 [len=0.439]
n348 -- n354 [len=0.439]
n348 -- n356 [len=0.439]
n348 -- n358 [len=0.439]
n348 -- n360 [len=0.439]
n348 -- n362 [len=0.439]
n348 -- n364 [len=0.439]
n348 -- n366 [len=0.439]
n349 -- n350 [len=0.367]
n351 -- n352 [len=0.367]
n351 -- n353 [len=0.367]
n354 -- n355 [len=0.367]
n356 -- n357 [len=0.367]
n358 -- n359 [len=0.367]
n360 -- n361 [len=0.367]
n362 -- n363 [len=0.367]
n364 -- n365 [len=0.367]
n366 -- n367 [len=0.367]
n368 -- n369 [len=0.439]
n370 -- n371 [len=0.439]
n370 -- n373 [len=0.439]
n370 -- n375 [len=0.439]
n371 -- n372 [len=0.367]
n373 -- n374 [len=0.367]
n375 -- n376 [len=0.367]
n377 -- n378 [len=0.439]
n379 -- n380 [len=0.439]
n379 -- n382 [len=0.439]
n379 -- n385 [len=0.439]
n379 -- n387 [len=0.439]
n379 -- n390 [len=0.439]
n379 -- n393 [len=0.439]
n379 -- n396 [len=0.439]
n379 -- n399 [len=0.439]
n379 -- n402 [len=0.439]
n379 -- n405 [len=0.439]
n380 -- n381 [len=0.367]
n382 -- n383 [len=0.367]
n383 -- n384 [len=0.294]
n385 -- n386 [len=0.367]
n387 -- n388 [len=0.367]
n388 -- n389 [len=0.294]
n390 -- n391 [len=0.367]
n391 -- n392 [len=0.294]
n393 -- n394 [len=0.367]
n394 -- n395 [len=0.294]
n396 -- n397 [len=0.367]
n397 -- n398 [len=0.294]
n399 -- n400 [len=0.367]
n400 -- n401 [len=0.294]
n402 -- n403 [len=0.367]
n403 -- n404 [len=0.294]
n405 -- n406 [len=0.367]
n406 -- n407 [len=0.294]
}")

networkD3
URL <- paste0(
"https://cdn.rawgit.com/christophergandrud/networkD3/",
"master/JSONdata/energy.json")
Energy <- jsonlite::fromJSON(URL)
# Plot
sankeyNetwork(Links = Energy$links, Nodes = Energy$nodes, Source = "source",
Target = "target", Value = "value", NodeID = "name",
units = "TWh", fontSize = 12, nodeWidth = 30)
Sankey diagrams
source: http://christophergandrud.github.io/networkD3/

library(networkD3)
library(networkD3)
data(MisLinks, MisNodes)
forceNetwork(Links = MisLinks, Nodes = MisNodes, Source = "source",
Target = "target", Value = "value", NodeID = "name",
Group = "group", opacity = 0.4)
source:http://christophergandrud.github.io/networkD3/
DiagrammeR -mermaid
mermaid("
+ sequenceDiagram
+ customer->>ticket seller: ask ticket
+ ticket seller->>database: seats
+ alt tickets available
+ database->>ticket seller: ok
+ ticket seller->>customer: confirm
+ customer->>ticket seller: ok
+ ticket seller->>database: book a seat
+ ticket seller->>printer: print ticket
+ else sold out
+ database->>ticket seller: none left
+ ticket seller->>customer: sorry
+ end
+ ")

library(sparkline)
load("C:/github/HtmlWidgetExamples/data/climate.RData")
library(data.table)
library(reshape2)
library(dplyr)
library(DT)
library(sparkline)
dat <- mutate(dat, Decade=paste0(Year - Year %% 10, "s"))
r <- range(filter(dat, Var=="Temperature")$Val)
# @knitr table
datatable(dat, rownames=FALSE)
# @knitr defs
colDefs1 <- list(list(targets=c(1:12), render=JS("function(data, type, full){ return '<span class=spark>' + data + '</span>' }")))
colDefs2 <- list(list(targets=c(1:6), render=JS("function(data, type, full){ return '<span class=spark>' + data + '</span>' }")))
# @knitr callbacks
bar_string <- "type: 'bar', barColor: 'orange', negBarColor: 'purple', highlightColor: 'black'"
cb_bar = JS(paste0("function (oSettings, json) { $('.spark:not(:has(canvas))').sparkline('html', { ", bar_string, " }); }"), collapse="")
line_string <- "type: 'line', lineColor: 'black', fillColor: '#ccc', highlightLineColor: 'orange', highlightSpotColor: 'orange'"
cb_line = JS(paste0("function (oSettings, json) { $('.spark:not(:has(canvas))').sparkline('html', { ", line_string, ", chartRangeMin: ", r[1], ", chartRangeMax: ", r[2], " }); }"), collapse="")
box_string <- "type: 'box', lineColor: 'black', whiskerColor: 'black', outlierFillColor: 'black', outlierLineColor: 'black', medianColor: 'black', boxFillColor: 'orange', boxLineColor: 'black'"
cb_box1 = JS(paste0("function (oSettings, json) { $('.spark:not(:has(canvas))').sparkline('html', { ", box_string," }); }"), collapse="")
cb_box2 = JS(paste0("function (oSettings, json) { $('.spark:not(:has(canvas))').sparkline('html', { ", box_string, ", chartRangeMin: ", r[1], ", chartRangeMax: ", r[2], " }); }"), collapse="")
# @knitr sparklines
dat.p <- filter(dat, Var=="Precipitation" & Decade=="2000s" & Month=="Aug")$Val
dat.p
# @knitr sparkline_dt_prep
dat.t <- filter(dat, Var=="Temperature") %>%
group_by(Decade, Month) %>% summarise(Temperature=paste(Val, collapse = ","))
dat.ta <- dcast(dat.t, Decade ~ Month)
dat.tb <- dcast(dat.t, Month ~ Decade)
# @knitr table_DxM_line
d1 <- datatable(data.table(dat.ta), rownames=FALSE, options=list(columnDefs=colDefs1, fnDrawCallback=cb_line))
d1$dependencies <- append(d1$dependencies, htmlwidgets:::getDependency('sparkline'))
d1
# @knitr table_MxD_bar
d2 <- datatable(data.table(dat.tb), rownames=FALSE, options=list(columnDefs=colDefs2, fnDrawCallback=cb_bar))
d2$dependencies <- append(d2$dependencies, htmlwidgets:::getDependency('sparkline'))
d2
# @knitr table_MxD_box1
d3 <- datatable(data.table(dat.tb), rownames=FALSE, options=list(columnDefs=colDefs2, fnDrawCallback=cb_box1))
d3$dependencies <- append(d3$dependencies, htmlwidgets:::getDependency('sparkline'))
d3
# @knitr table_MxD_box2
d4 <- datatable(data.table(dat.tb), rownames=FALSE, options=list(columnDefs=colDefs2, fnDrawCallback=cb_box2))
d4$dependencies <- append(d4$dependencies, htmlwidgets:::getDependency('sparkline'))
d4
# @knitr final_prep
dat.t2 <- filter(dat, Var=="Temperature" & Month=="Aug") %>%
group_by(Location, Month, Var, Decade) %>%
summarise(Mean=round(mean(Val), 1), SD=round(sd(Val), 2), Min=min(Val), Max=max(Val), Samples=paste(Val, collapse = ",")) %>%
mutate(Series=Samples) %>% data.table
cd <- list(list(targets=8, render=JS("function(data, type, full){ return '<span class=sparkSamples>' + data + '</span>' }")),
list(targets=9, render=JS("function(data, type, full){ return '<span class=sparkSeries>' + data + '</span>' }")))
cb = JS(paste0("function (oSettings, json) {
$('.sparkSeries:not(:has(canvas))').sparkline('html', { ", line_string, " });
$('.sparkSamples:not(:has(canvas))').sparkline('html', { ", box_string, " });
}"), collapse="")
# @knitr table_final
d5 <- datatable(data.table(dat.t2), rownames=FALSE, options=list(columnDefs=cd, fnDrawCallback=cb))
d5$dependencies <- append(d5$dependencies, htmlwidgets:::getDependency('sparkline'))
d5

markov chains in R
> ggplot(all_mod_plot, aes(x = conv_type, y = value, group = channel_name)) +
+ theme_solarized(base_size = 18, base_family = "", light = TRUE) +
+ scale_color_manual(values = pal(10)) +
+ scale_fill_manual(values = pal(10)) +
+ geom_line(aes(color = channel_name), size = 2.5, alpha = 0.8) +
+ geom_point(aes(color = channel_name), size = 5) +
+ geom_label_repel(aes(label = paste0(channel_name, ': ', value), fill = factor(channel_name)),
+ alpha = 0.7,
+ fontface = 'bold', color = 'white', size = 5,
+ box.padding = unit(0.25, 'lines'), point.padding = unit(0.5, 'lines'),
+ max.iter = 100) +
+ theme(legend.position = 'none',
+ legend.title = element_text(size = 16, color = 'black'),
+ legend.text = element_text(size = 16, vjust = 2, color = 'black'),
+ plot.title = element_text(size = 20, face = "bold", vjust = 2, color = 'black', lineheight = 0.8),
+ axis.title.x = element_text(size = 24, face = "bold"),
+ axis.title.y = element_text(size = 16, face = "bold"),
+ axis.text.x = element_text(size = 16, face = "bold", color = 'black'),
+ axis.text.y = element_blank(),
+ axis.ticks.x = element_blank(),
+ axis.ticks.y = element_blank(),
+ panel.border = element_blank(),
+ panel.grid.major = element_line(colour = "grey", linetype = "dotted"),
+ panel.grid.minor = element_blank(),
+ strip.text = element_text(size = 16, hjust = 0.5, vjust = 0.5, face = "bold", color = 'black'),
+ strip.background = element_rect(fill = "#f0b35f")) +
+ labs(x = 'Model', y = 'Conversions') +
+ ggtitle('Models comparison') +
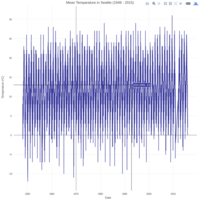
library(plotly)
library(plotly)
# Read some weather data
df <- read.csv('https://cdn.rawgit.com/plotly/documentation/source/_posts/r/scattergl/weather-data.csv')
# Convert to dates
df$Date <- zoo::as.Date(df$Date, format = "%m/%d/%Y")
p <- plot_ly(df, x = Date, y = Mean_TemperatureC, name = "Mean Temp.", type = "scattergl",
marker = list(color = "#3b3b9e")) %>%
layout(title = "Mean Temparature in Seattle (1948 - 2015)",
yaxis = list(title = "Temperature (<sup>o</sup>C)"))
p

plotly example
library(plotly)
p <- plot_ly(plotly::mic, r = r, t = t, color = nms, mode = "lines")
layout(p, title = "Mic Patterns", orientation = -90)
plotly...map.
df <- read.csv('https://raw.githubusercontent.com/plotly/datasets/master/globe_contours.csv')
df$id <- seq_len(nrow(df))
library(tidyr)
d <- df %>%
gather(key, value, -id) %>%
separate(key, c("l", "line"), "\\.") %>%
spread(l, value)
p <- plot_ly(type = 'scattergeo', mode = 'lines',
line = list(width = 2, color = 'violet'))
for (i in unique(d$line))
p <- add_trace(p, lat = lat, lon = lon, data = subset(d, line == i))
geo <- list(
showland = TRUE,
showlakes = TRUE,
showcountries = TRUE,
showocean = TRUE,
countrywidth = 0.5,
landcolor = toRGB("grey90"),
lakecolor = toRGB("white"),
oceancolor = toRGB("white"),
projection = list(
type = 'orthographic',
rotation = list(
lon = -100,
lat = 40,
roll = 0
)
),
lonaxis = list(
showgrid = TRUE,
gridcolor = toRGB("gray40"),
gridwidth = 0.5
),
lataxis = list(
showgrid = TRUE,
gridcolor = toRGB("gray40"),
gridwidth = 0.5
)
)
layout(p, showlegend = FALSE, geo = geo,
title = 'Contour lines over globe<br>(Click and drag to rotate)')
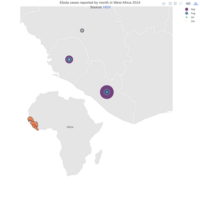
plotly example
library(plotly)
df <- read.csv('https://raw.githubusercontent.com/plotly/datasets/master/2014_ebola.csv')
# restrict from June to September
df <- subset(df, Month %in% 6:9)
# ordered factor variable with month abbreviations
df$abbrev <- ordered(month.abb[df$Month], levels = month.abb[6:9])
# September totals
df9 <- subset(df, Month == 9)
# common plot options
g <- list(
scope = 'africa',
showframe = F,
showland = T,
landcolor = toRGB("grey90")
)
# styling for "zoomed in" map
g1 <- c(
g,
resolution = 50,
showcoastlines = T,
countrycolor = toRGB("white"),
coastlinecolor = toRGB("white"),
projection = list(type = 'Mercator'),
list(lonaxis = list(range = c(-15, -5))),
list(lataxis = list(range = c(0, 12))),
list(domain = list(x = c(0, 1), y = c(0, 1)))
)
g2 <- c(
g,
showcountries = F,
bgcolor = toRGB("white", alpha = 0),
list(domain = list(x = c(0, .6), y = c(0, .6)))
)
plot_ly(df, type = 'scattergeo', mode = 'markers', locations = Country,
locationmode = 'country names', text = paste(Value, "cases"),
color = as.ordered(abbrev), marker = list(size = Value/50), inherit = F) %>%
add_trace(type = 'scattergeo', mode = 'text', geo = 'geo2', showlegend = F,
# plotly should support "unboxed" constants
lon = list(21.0936), lat = list(7.1881), text = list('Africa')) %>%
add_trace(type = 'choropleth', locations = Country, locationmode = 'country names',
z = Month, colors = "black", showscale = F, geo = 'geo2', data = df9) %>%
layout(title = 'Ebola cases reported by month in West Africa 2014<br> Source: <a href="https://data.hdx.rwlabs.org/dataset/rowca-ebola-cases">HDX</a>',
geo = g1, geo2 = g2)
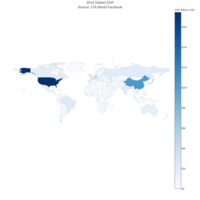
plotly example
df <- read.csv('https://raw.githubusercontent.com/plotly/datasets/master/2014_world_gdp_with_codes.csv')
# light grey boundaries
l <- list(
color = toRGB("grey"),
width = 0.5
)
# specify map projection/options
g <- list(
showframe = FALSE,
showcoastlines = FALSE,
projection = list(type = 'Mercator')
)
plot_ly(df, z = GDP..BILLIONS., text = COUNTRY, locations = CODE, type = 'choropleth',
color = GDP..BILLIONS., colors = 'Blues', marker = list(line = l),
colorbar = list(tickprefix = '$', title = 'GDP Billions US$')) %>%
# TODO: how to add the hyperlink? (<a href=""> doesn't seem to work)
layout(title = '2014 Global GDP<br>Source: CIA World Factbook', geo = g)

plotly example
df <- read.csv("https://raw.githubusercontent.com/plotly/datasets/master/2011_us_ag_exports.csv")
df$hover <- with(df, paste(state, '<br>', "Beef", beef, "Dairy", dairy, "<br>",
"Fruits", total.fruits, "Veggies", total.veggies,
"<br>", "Wheat", wheat, "Corn", corn))
# give state boundaries a white border
l <- list(
color = toRGB("white"),
width = 2
)
# specify some map projection/options
g <- list(
scope = 'usa',
projection = list(type = 'albers usa'),
showlakes = TRUE,
lakecolor = toRGB('white')
)
plot_ly(df, z = total.exports, text = hover, locations = code, type = 'choropleth',
locationmode = 'USA-states', color = total.exports, colors = 'Purples',
marker = list(line = l)), colorbar = list(title = "Millions USD")) %>%
layout(title = '2011 US Agriculture Exports by State<br>(Hover for breakdown)', geo = g)

plotly example
p <- plot_ly(plotly::mic, r = r, t = t, color = nms, mode = "lines")
layout(p, title = "Mic Patterns", orientation = -90)
p <- plot_ly(plotly::hobbs, r = r, t = t, color = nms, opacity = 0.7, mode = "markers")
layout(p, title = "Hobbs-Pearson Trials", plot_bgcolor = toRGB("grey90"))
p <- plot_ly(plotly::wind, r = r, t = t, color = nms, type = "area")
layout(p, radialaxis = list(ticksuffix = "%"), orientation = 270)
plotly
library(plotly)
plot_ly(z = volcano, type = "contour")
#' Advanced
x <- rnorm(200)
y <- rnorm(200)
p1 <- plot_ly(x = x, type = "histogram")
p2 <- plot_ly(x = x, y = y, type = "histogram2dcontour")
p3 <- plot_ly(y = y, type = "histogram")
a1 <- list(domain = c(0, .85))
a2 <- list(domain = c(.85, 1))
subplot(
layout(p1, xaxis = a1, yaxis = a2),
layout(p2, xaxis = a1, yaxis = a1),
layout(p3, xaxis = a2, yaxis = a1)
)

plotly example
library(plotly)
#' basic boxplot
plot_ly(y = rnorm(50), type = "box") %>%
add_trace(y = rnorm(50, 1))
#' adding jittered points
plot_ly(y = rnorm(50), type = "box", boxpoints = "all", jitter = 0.3,
pointpos = -1.8)
#' several box plots
data(diamonds, package = "ggplot2")
plot_ly(diamonds, y = price, color = cut, type = "box")
#' grouped box plots
plot_ly(diamonds, x = cut, y = price, color = clarity, type = "box") %>%
layout(boxmode = "group")
plotly
library(dplyr)
ggplot2::diamonds %>% count(cut) %>%
plot_ly(x = cut, y = n, type = "bar", marker = list(color = toRGB("black")))
# mapping a color variable
ggplot2::diamonds %>% count(cut, clarity) %>%
plot_ly(x = cut, y = n, type = "bar", color = clarity)

Publish HTML
s <- matrix(c(1, .5, .5,
.5, 1, .5,
.5, .5, 1), ncol = 3)
# use the mvtnorm package to sample 200 observations
obs <- mvtnorm::rmvnorm(200, sigma = s)
# collect everything in a data-frame
df <- setNames(data.frame(obs), c("x", "y", "z"))
library(plotly)
plot_ly(df, x = x, y = y, z = z, type = "scatter3d", mode = "markers")
plotly
library(plotly)
p <- plot_ly(midwest, x = percollege, color = state, type = "box")
p
DiagrammeR
grViz("digraph {layout = twopi
+ node [shape = circle]
+ V -> {O L K A N}}")

higncharter viridislite treemap flexdashboard
https://beta.rstudioconnect.com/jjallaire/htmlwidgets-highcharter/
library(highcharter)
library(dplyr)
library(viridisLite)
library(forecast)
library(treemap)
library(flexdashboard)
thm <-
hc_theme(
colors = c("#1a6ecc", "#434348", "#90ed7d"),
chart = list(
backgroundColor = "transparent",
style = list(fontFamily = "Source Sans Pro")
),
xAxis = list(
gridLineWidth = 1
)
)
```
Column {data-width=600}
-----------------------------------------------------------------------
### Sales Forecast
```{r}
AirPassengers %>%
forecast(level = 90) %>%
hchart() %>%
hc_add_theme(thm)
```
### Sales by State
```{r}
data("USArrests", package = "datasets")
data("usgeojson")
USArrests <- USArrests %>%
mutate(state = rownames(.))
n <- 4
colstops <- data.frame(
q = 0:n/n,
c = substring(viridis(n + 1), 0, 7)) %>%
list.parse2()
highchart() %>%
hc_add_series_map(usgeojson, USArrests, name = "Sales",
value = "Murder", joinBy = c("woename", "state"),
dataLabels = list(enabled = TRUE,
format = '{point.properties.postalcode}')) %>%
hc_colorAxis(stops = colstops) %>%
hc_legend(valueDecimals = 0, valueSuffix = "%") %>%
hc_mapNavigation(enabled = TRUE) %>%
hc_add_theme(thm)
```
Column {.tabset data-width=400}
-----------------------------------------------------------------------
### Sales by Category
```{r, fig.keep='none'}
data("Groceries", package = "arules")
dfitems <- tbl_df(Groceries@itemInfo)
set.seed(10)
dfitemsg <- dfitems %>%
mutate(category = gsub(" ", "-", level1),
subcategory = gsub(" ", "-", level2)) %>%
group_by(category, subcategory) %>%
summarise(sales = n() ^ 3 ) %>%
ungroup() %>%
sample_n(31)
tm <- treemap(dfitemsg, index = c("category", "subcategory"),
vSize = "sales", vColor = "sales",
type = "value", palette = rev(viridis(6)))
highchart() %>%
hc_add_series_treemap(tm, allowDrillToNode = TRUE,
layoutAlgorithm = "squarified") %>%
hc_add_theme(thm)
```
### Best Sellers
```{r}
set.seed(2)
nprods <- 10
dfitems %>%
sample_n(nprods) %>%
.$labels %>%
rep(times = sort(sample( 1e4:2e4, size = nprods), decreasing = TRUE)) %>%
factor(levels = unique(.)) %>%
hchart(showInLegend = FALSE, name = "Sales", pointWidth = 10) %>%
hc_add_theme(thm) %>%
hc_chart(type = "bar")

ggplot2- shiny-plot
https://beta.rstudioconnect.com/jjallaire/htmlwidgets-highcharter/

correlogram-corrgram
require(corrgram)
> corrgram(iris)

paleofire R
all_sites<-pfSiteSel()
> plot(all_sites)

ggbiplot
> library(ggbiplot)
> data(wine)
> wine.pca <- prcomp(wine, scale. = TRUE)
> g <- ggbiplot(wine.pca, obs.scale = 1, var.scale = 1,
+ groups = wine.class, ellipse = TRUE, circle = TRUE)
> g <- g + scale_color_discrete(name = '')
> g <- g + theme(legend.direction = 'horizontal',
+ legend.position = 'top')
> print(g)

parcoord
library(MASS)
> parcoord(iris[1:4], col = iris$Species)

heatmap in R
dist.dist.matrix <- as.matrix(dist(iris[, 1:4]))
> heatmap(dist.dist.matrix)

scatterplot3d
library(scatterplot3d)
> scatterplot3d(iris$Petal.Width, iris$Sepal.Length, iris$Sepal.Width)
> scatterplot3d(iris$Petal.Width, iris$Sepal.Length, iris$Sepal.Width)